Figure 4.
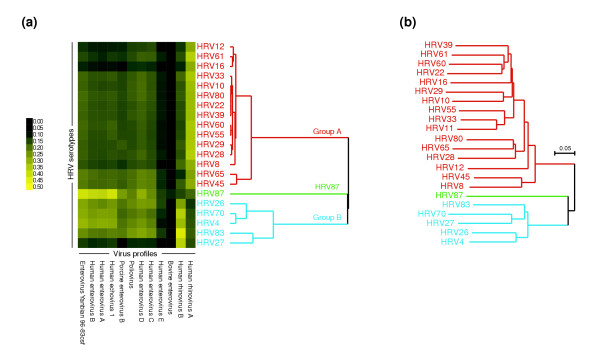
Human rhinovirus (HRV) serotype discrimination using E-Predict similarity scores. (a) Culture samples of 22 distinct HRV serotypes were separately hybridized to the microarray. E-Predict similarity scores were obtained for all virus profiles in the energy matrix and clustered using average linkage hierarchical clustering and Pearson correlation as the similarity metric. Virus profiles for which similarity scores could be calculated in all 22 experiments were included in the clustering. Both microarrays (rows) and virus profiles (columns) were clustered. (b) Published nucleotide sequences of VP1 capsid protein from the 22 HRV serotypes were aligned using ClustalX. Phylogenetic tree based on the resulting alignment is shown.
