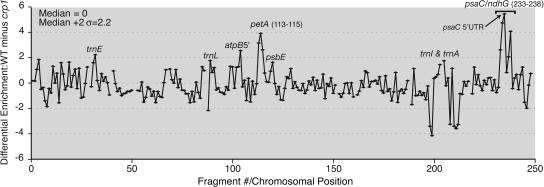
Figure 5.
CRP1 RIP-Chip Data Plotted According to Genomic Position.
The log2-transformed enrichment ratios (F635:F532) were normalized between the three CRP1 RIP-chip assays involving wild-type stroma and the two control assays with crp1 mutant stroma. The difference between the median normalized values for replicate spots in the experimental and control data sets is plotted as a function of fragment number. Fragments for which fewer than two spots per array yielded an F532 signal above background were excluded and appear as gaps in the curve. The data used to generate this curve are provided in Supplemental Table 3 online. Supplemental Figure 3 online shows analogous plots for the five individual experiments without data normalization. The valley within the psaC peak region is artifactual: the F532 signal/base pair for fragment 236 (the low point) is substantially lower than that for the fragments that overlap it, indicating that the DNA in this spot is defective.