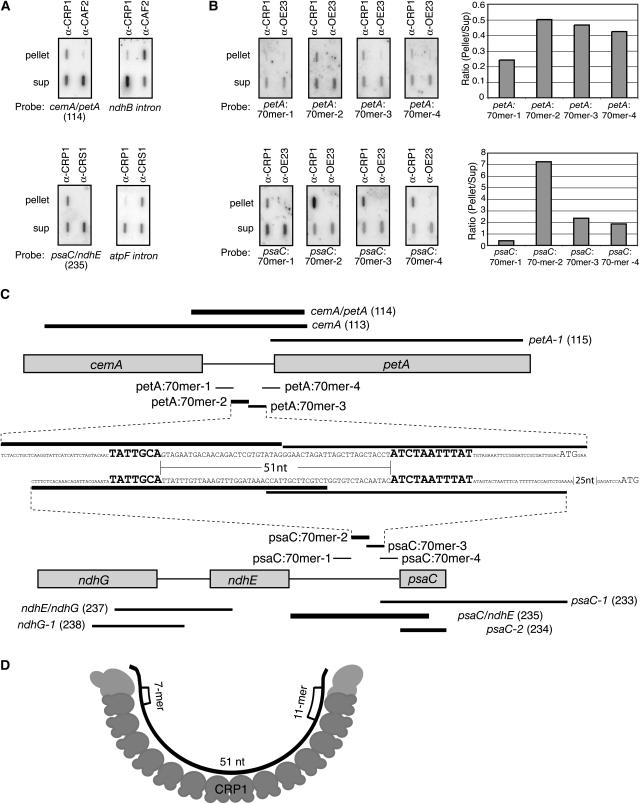
Figure 6.
Fine-Mapping of RNAs That Coimmunoprecipitate with CRP1.
(A) Verification that petA and psaC RNAs are enriched in CRP1 coimmunoprecipitates. Coimmunoprecipitations and RNA extractions were performed as for the RIP-chip assays; the RNAs were then analyzed by slot-blot hybridization with the indicated probes. The top panels, showing CRP1-dependent enrichment of petA 5′ sequences, are duplicate blots of RNAs from parallel CRP1 and CAF2 immunoprecipitations. The CAF2 immunoprecipitation was included as a negative control; the ndhB intron is a known CAF2 substrate (Ostheimer et al., 2003) and was analyzed to demonstrate the integrity of the CAF2 pellet RNA. The bottom panels, showing CRP1-dependent enrichment of psaC 5′ sequences, are duplicate blots of RNAs from parallel CRP1 and CRS1 immunoprecipitations. The CRS1 immunoprecipitation was included as a negative control; the atpF intron is a known CRS1 substrate (Ostheimer et al., 2003) and was analyzed to demonstrate the integrity of the CRS1 pellet RNA. The psaC/ndhE (fragment 235) and cemA/petA (fragment 114) probes correspond to the two highest ranking fragments in the CRP1 RIP-chip experiments and are diagrammed in (C). Sup, supernatant.
(B) Fine-mapping of CRP1-associated RNAs from the psaC and petA 5′ UTRs. Immunoprecipitation with a different antibody (α-OE23) served as a negative control. Duplicate slot blots were prepared as in (A) except that RNase inhibitor was omitted from the immunoprecipitation reactions. Blots were probed with 5′ end-labeled 70-mer oligonucleotides as diagrammed in (C). Results were quantified with a phosphorimager and plotted at right.
(C) Summary of CRP1 RIP-chip and slot-blot data for the psaC and petA regions. The PCR fragments on the microarray (identified by name and number) and the 70-mer oligonucleotides used for slot-blot probing are indicated with lines whose thickness reflects the degree to which the sequences were enriched in CRP1 immunoprecipitates. The nucleotide (nt) sequences shown span the pair of 70-mers from each region that detected the most highly enriched sequences; they are aligned to highlight the 7-mer and 11-mer motifs (large boldface letters), with start codons shown in large lightface letters.
(D) Model for RNA recognition by CRP1. The cartoon illustrates how a CRP1 monomer might bind the RNA encompassed by the shared 7-mer and 11-mer motifs in the psaC and petA 5′ UTRs. The 14 repeating units in CRP1 represent its 14 tandem PPR motifs. The RNA is shown bound to the concave face of the predicted PPR superhelix; this predicted surface of CRP1 appears well-suited for RNA binding (Williams and Barkan, 2003).