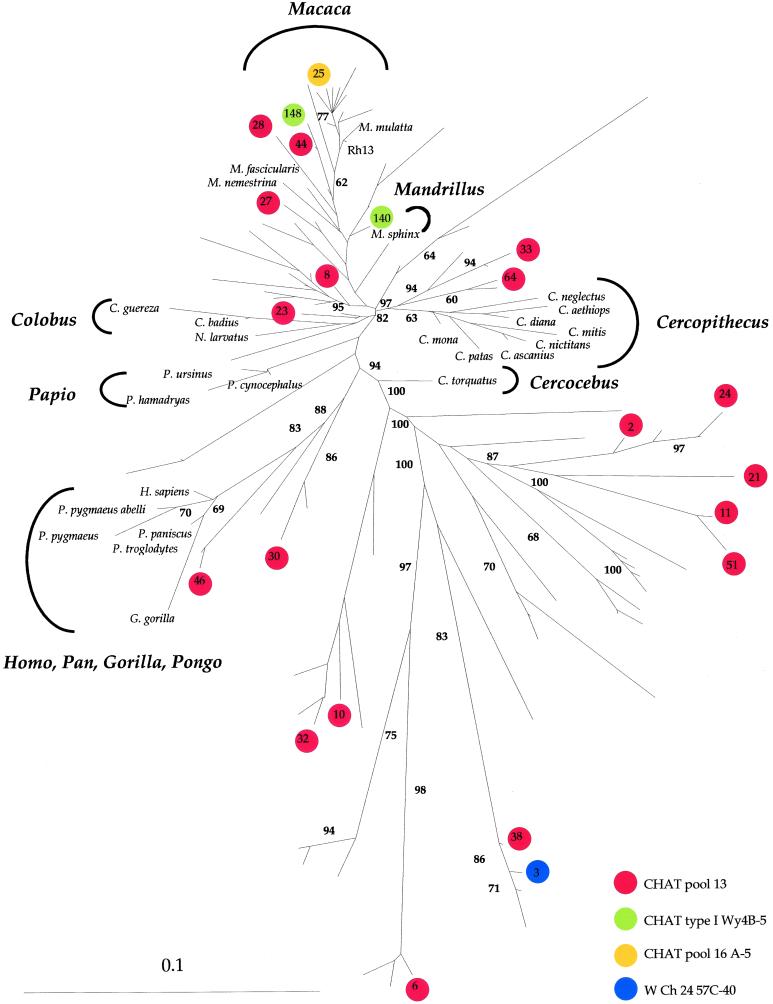
Figure 2.
Phylogenetic tree of a wide collection of 12S rDNA sequences. They include the numts from a single Chinese rhesus macaque (M. mulatta), 26 mtDNA sequences corresponding to primate lineages for reference, and 22 putative numts from OPV samples (22). The latter are indicated by colored circles, with each color corresponding to a different vaccine lot. The numbers refer to distinct numt clones. M. mulatta refers to the normal mtDNA haplotype of the Chinese rhesus macaque. Rh13 corresponds to the normal rhesus macaque allele from CHAT pool 13. Sequences were aligned by CLUSTAL W. The tree was derived by neighbor-joining analysis applied to pairwise sequence distances calculated with DNADIST of the PHYLIP 3.5 package using the Kimura two-parameter method to generate unrooted trees. Horizontal branch lengths are drawn to scale with the bar indicating 0.1 nucleotide replacements per site. The final output was generated with TREEVIEW. The number at each node represents the percentage of bootstrap replicates (out of 100). Only bootstrap values of ≥60 are given.