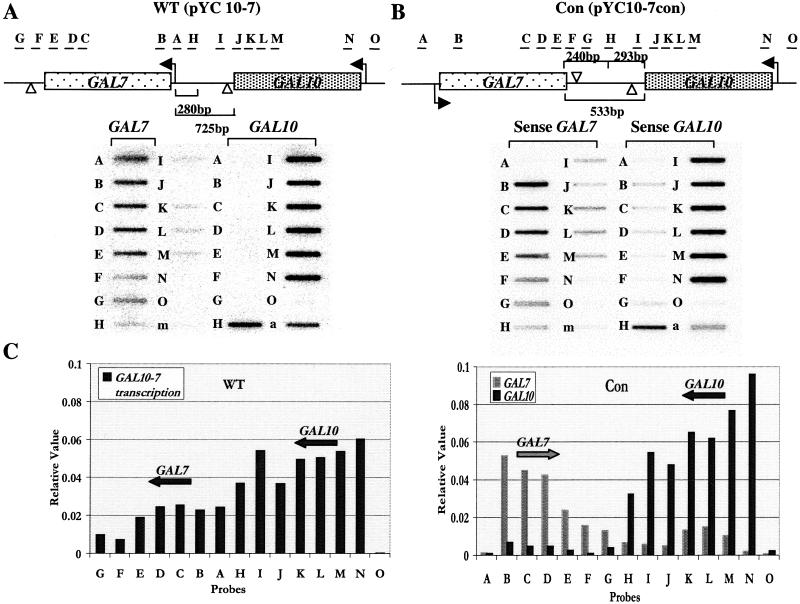
Figure 1.
TRO analysis of transformed GAL10-7 plasmids. (A) TRO analysis of pYC10-7 (WT) transformed cells. The diagram shows the tandem arrangement of the GAL10 and GAL7 genes, with the direction of transcription indicated with an arrow. The position of the single-stranded M13 probes used are shown and are as described in Table 1. Probe m is an M13 probe without an insert serving as a negative hybridization control. Probe a is an M13 probe with a fragment of the S. cerevisiae actin gene serving as a positive transcription control. The distances between the ORFs of GAL10 and GAL7 in the WT orientation are labeled. Open arrowheads indicate polyadenylation sites. The GAL7 promoter region is indicated also. (B) TRO analysis of pYC10-7Con (Con)-transformed cells. The diagram shows convergent orientation of GAL10 and GAL7, with the intergenic region's distance labeled. This construct includes 240 bp downstream of GAL7 and 293 bp downstream of GAL10 to make a hybrid intergenic region. Sense and antisense single-stranded M13 DNA probes were selected as appropriate. (C) Quantitation of the data shown in A and B. Signals were quantified in a PhosphorImager (Molecular Dynamics). The values were corrected for background hybridization and U content in each probe. Signals were normalized to the actin positive transcription probe a on each filter to allow direct comparison of transcription levels.