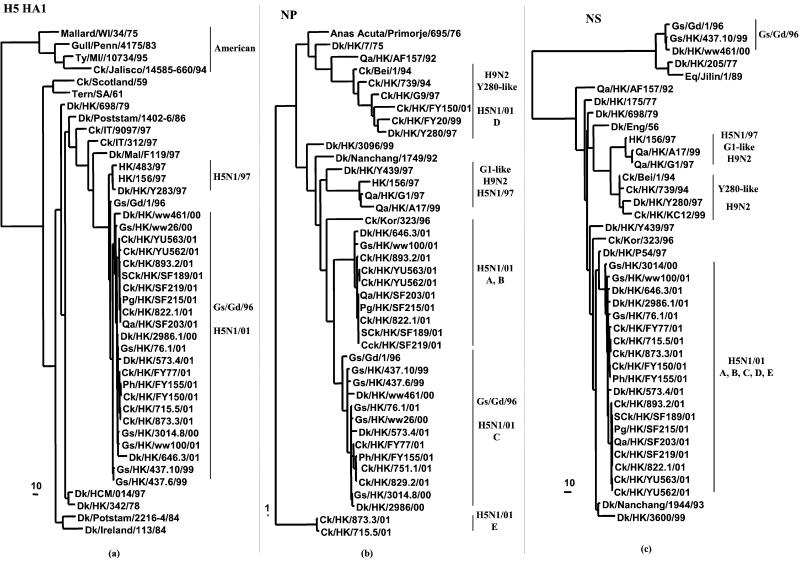
Figure 1.
Phylogenetic trees for the H5 HA1 (a), NP (b), and NS (c) genes of influenza A viruses. The nucleotide sequences were analyzed with PAUP by using a maximum-parsimony algorithm. Nucleotides 43 to 1,023 (981 bp) of the HA gene, 42 to 1,002 (961 bp) of the NP gene, and 41 to 824 (784 bp) of the NS gene were used for the phylogenetic analysis. The H5 HA1 phylogenetic tree is rooted to A/Japan/305+/57 (H2N2). The nucleotide trees of NP and NS gene are rooted to A/Equine/Prague/1/56 (H7N7). The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Names and abbreviations of viruses studied are listed in Table 1, and the abbreviations are denoted in the footnote. Other sequences can be found in GenBank.