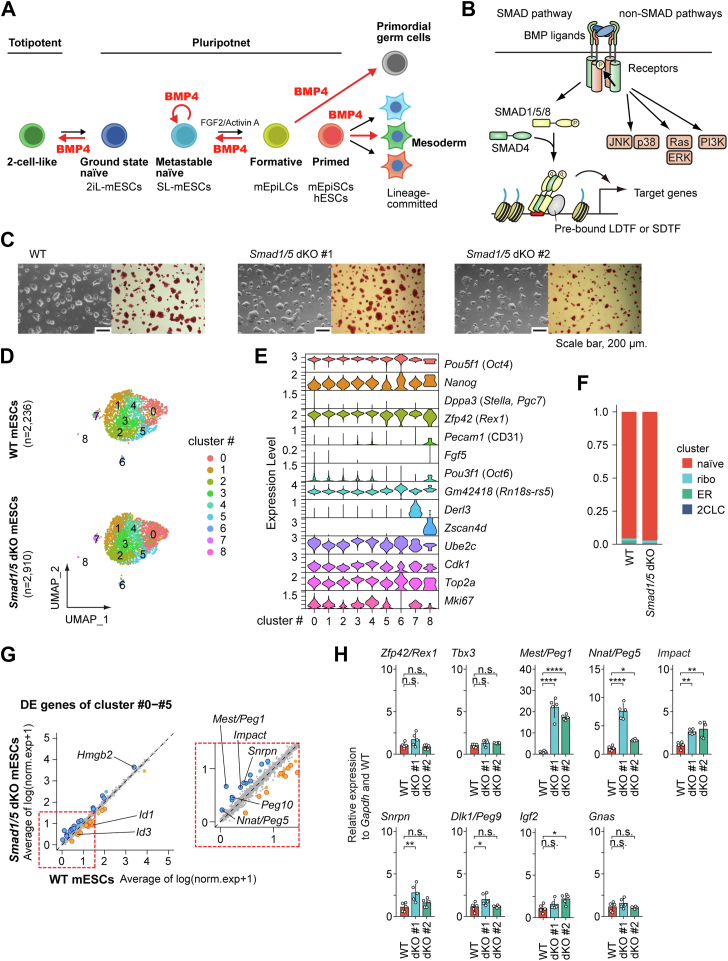
Figure 1.
Single-cell RNA-seq analysis of Smad1/5-deficient mESCs and parental mESCs. A, a schematic illustration of roles of BMP signaling in the biology of pluripotent stem cells (PSCs). B, a schematic illustration of signaling transduction of BMP signaling. C, morphology (left) and alkaline phosphatase staining (right) of S1/5 dKO cell lines in LIF + serum. Scale bar represents 200 μm. D, uniform manifold approximation and projection (UMAP) visualization of the transcriptional heterogeneity of parental E14 mESCs (WT, n = 2236) and Smad1/5-deficient mESCs (S1/5 dKO, n = 2910). Cells are colored based on the clusters they belong to. E, stacked violin plots of the integrated data showing the expression profile of marker genes: pluripotency (Pou5f1/OCT4), naïve pluripotency (Nanog, Zfp42), formative/primed pluripotency (Fgf5, Pou3f1/OCT6), late 2-cell stage (Zscan4d), and G2/M phase (Ube2c, Cdk1, Top2a). F, stacked bar plot showing the fraction of cell subtypes determined by marker gene expression. G, scatter plots showing expression patterns of genes in WT and S1/5 dKO cells. Average expression of log-normalized genes, difference between the transcriptomic profiles of WT and S1/5 dKO cells. Among the differentially expressed genes (DEGs), top 40 genes (20 upregulated and 20 downregulated) are highlighted in orange and light blue, respectively. In addition, those with SMAD1/5 binding within 10 kb from their transcription start sites (TSSs) are represented as rimmed circles. Orange: WT > S1/5 dKO genes, light blue: WT < S1/5 dKO genes. H, qRT–PCR analysis of mRNA of indicated genes in WT and S1/5 dKO cells. Gapdh was used as an endogenous control. Results of n = 5 independent biological replicates are presented as scatter plots with bar graphs, which indicate mean ± SD. Differences between the conditions were analyzed by Tukey's honestly significant difference test corrected for multiple comparisons. ∗, ∗∗, ∗∗∗, ∗∗∗∗, and ns represent p < 0.05, 0.01, 0.001, 0.0001, and not significant, respectively. BMP, bone morphogeneic protein; dKO, double KO; EpiLC, mouse epiblast stem cell-like cell; EpiSC, mouse epiblast stem cell; hESC, human embryonic stem cell; LDTF, lineage-determining transcription factor; LIF, leukemia inhibitory factor; mESC, mouse embryonic stem cell; P, phosphorylation; qRT–PCR, quantitative RT–PCR; SDTF, signal-dependent transcription factor.