Abstract
The role of the hepatitis B virus X protein (HBx) in the pathogenesis of hepatitis B virus (HBV) infection remains unclear. HBx exhibits pleiotropic biological effects, whose in vivo relevance is a matter for debate. In the present report, we have used a combination of HBx-expressing transgenic mice and liver cell transplantation to investigate the in vivo impact of HBx expression on liver cell proliferation and viability in a regenerative context. We show that moderate HBx expression inhibits liver regeneration after partial hepatectomy in HBx-expressing transgenic mice. We also demonstrate that the transplantation of HBx-expressing liver cells, isolated from HBx transgenic mice, is sufficient to inhibit overall recipient liver regeneration after partial hepatectomy. Moreover, the injection of serum samples drawn from HBx-expressing transgenic mice mimicked the inhibitory effect of HBx on liver regeneration. Finally, the incubation of primary rat hepatocytes with the supernatant of HBx-expressing liver cells inhibits cellular DNA synthesis. Taken together, our results demonstrate a paracrine inhibitory effect of HBx on liver cell proliferation and lead us to propose HBV as one of the few viruses implicated in human cancer which act, at least in part, through paracrine biological pathways.
Hepatitis B virus (HBV) is a major pathogen that chronically infects more than 300 million individuals worldwide. Chronic HBV infection is an important cause of death through induction of cirrhosis and hepatocellular carcinoma (HCC) (1–3). Liver inflammation triggered by the host immune response to the virus is a major factor contributing to the development of HCC; it induces fibrosis and liver cell proliferation and, eventually, deregulated liver cell expansion (4, 5). There is also evidence that HBV has a direct impact on the cellular machinery. Integration of the viral genome into the host genome can induce chromosomal instability; it may also integrate in the vicinity of genes controlling cell proliferation, viability, and differentiation. Our recent findings have shown that, in contrast with previously held views, this mechanism may in fact be prevalent (1, 6, 7).
In addition, a number of studies have shown the capacity of certain HBV proteins, namely HBVx protein (HBx) (8–10), truncated envelope PreS2/St (11), and a splice-generated protein (HBSP) (12), to modulate cell proliferation and viability. HBx has, in particular, been the focus of several investigations. This protein is expressed at all stages of viral infection and has been implicated in the establishment of viral infection in woodchucks infected by the woodchuck hepatitis virus (WHV), an HBV-related virus from the Hepadna virus family (13). In most in vitro and in vivo studies, HBx has exhibited no, or only a very weak, oncogenic effect, although HBx has been shown to induce HCC in certain lines of transgenic mice (14) and may cooperate in vitro, under some conditions, with activated ras oncogene (15). Taken together, these observations and the overall results obtained in various HBx-expressing transgenic mice with different genetic backgrounds have led to the suggestion that HBx may act as a promoting agent during liver carcinogenesis, in particular, by cooperating with the c-myc oncogene and by sensitizing liver cells to carcinogenic chemicals (16, 17).
HBx exhibits a predominantly cytoplasmic localization (partly associated with the proteasome and mitochondria) although, at least under some experimental conditions, nuclear HBx can be also detected (18–20). In vitro, it has been well established that HBx transactivates a variety of cellular gene promoters by acting on several cis-acting elements such as NF-κB, AP1 and AP2, and CRE (8, 10). HBx does not bind DNA. It activates signal-transduction cascades such as the Ras/Raf mitogen-activated protein kinase, c-Jun NH2-terminal kinase, JAK/STAT, and, possibly, protein kinase C (21–24). HBx may also directly interact with transcription factors, such as cAMP response element binding protein/activating transcription factor (CREB/ATF) (8), and with regulatory molecules, such as IκBα (25), it may also affect basal transcriptional regulation by binding RNA polymerase II subunit 5 (26), transcription factor IIB (27, 28), TATA-binding proteins, and transcription factor II (26, 29). In addition, HBx may also directly bind a number of important cellular regulatory molecules controlling cell viability and proliferation, such as p53 (30), UVDD (31), ERCC3 DNA repair (32), nuclear export CRM1 (33), and senescence-related factor (34).
Depending on the experimental conditions, discrepant results have been shown regarding the effects of HBx ectopic expression on cell proliferation and apoptosis. In fact, HBx has been reported to block p53- (35), Fas-, tumor necrosis factor α (TNFα)- (36), transforming growth factor β (TGFβ)- (37) dependent apoptosis as well as caspase 3 activity (38); in most recent reports, however, HBx has been shown to induce or sensitize to apoptosis, by using p53-dependent and independent mechanisms (39–44). HBx-dependent apoptosis can be suppressed by activating the phosphatidylinositol 3 kinase/Akt (45), insulin-like growth factor II (43), and bcl2 pathways (42, 46). The level of HBx expression achieved in experiments based mainly on transient expression and the cell type investigated is an important factor influencing the overall cellular effects of HBx; the importance of the intracellular level of HBx has been well illustrated in in vivo experiments with HBx-expressing transgenic mice (16, 41, 47, 48). In addition, our group and others have recently shown that mutations identified in HBx-encoding sequences in the tumor cells of HCC patients can markedly modify the biological activity of HBx and abrogate its proapoptotic effects. Our results have led us to suggest a model, whereby mutations in HBx sequences may favor the selection and, thus, the clonal expansion of certain HBV-infected liver cells in chronically infected HBV carriers (9, 44, 49).
As mentioned above, studies based on the in vitro ectopic expression of HBx produced variable results that reflect the different in vitro experimental protocols used. Extracellular signals may also significantly affect the differentiation status of a cell and its response to HBx expression; in this respect, in vivo studies in HBx-expressing transgenic mice have provided important information but their results have also shown discrepancies, due in particular to the different genetic backgrounds of the mice used and the different levels of HBx accumulation (14, 16, 50). In addition, the expression of HBx in most transgenic mouse liver cells does not fully mimic the situation observed in infected patients. Indeed, HBx is expressed in a small proportion of liver cells in patients with chronic hepatitis and cirrhosis and in the tumor cells of patients with HCC (19, 49, 51). Moreover, a major feature of the pathogenesis of HBV infection is the expression of viral proteins in the context of inflammation and liver cell regeneration (4).
In the present study, we wished to address the major issue of the in vivo effects of HBx on cell proliferation and viability in the context of a regenerative signal. Therefore, using HBx-expressing transgenic mice, we first investigated whether HBx expression affected the potential for liver regeneration after partial hepatectomy. We took advantage of the interesting characteristics of the AX16, HBx-expressing, transgenic line (16, 52), that expresses a moderate amount of HBx soon after birth, the levels declining as the animals become older. Furthermore, we benefited from recent advances in liver cell transplantation, which may offer new experimental approaches to address these issues (53–55). We reasoned that the transplantation of HBx-expressing hepatocytes isolated from HBx-expressing transgenic mice into nontransgenic mouse liver, and the induction of liver regeneration by partial hepatectomy would indeed allow us to investigate the effects of HBx expression in the context of a low percentage of HBx-expressing liver cells undergoing in vivo proliferation. Using this approach, we demonstrate a paracrine action of HBx and propose a model for HBx-dependent pathogenesis.
Materials and Methods
Animals.
AX16 transgenic mice carry the HBx ORF linked to the promoter-enhancer of the human antithrombin III gene and have been described in detail elsewhere (52). The HNF1-LacZ transgenic mice carry the nls-LacZ gene driven by the regulatory region of the hepatocyte nuclear factor 1 (HNF-1) rat gene. The transgene contains 3.3 kb of the HNF-1 promoter (56) linked to the nls-LacZ gene containing the simian virus 40 early region polyadenylation signal (57) (gift from M. Pontoglio and M. Yaniv, Pasteur Institute, Paris). AX16/HNF1-LacZ double-transgenic mice were generated by crossing homozygous AX16 and HNF1-LacZ mice, derived from the same parental strain of inbred mice. The experiments were then carried out on these AX16/HNF1-LacZ double-transgenic mice (referred to as HBx-LacZ) and using HNF1-LacZ transgenic mice (referred to as LacZ) as controls. Previous studies have documented, by using Western Blot analysis, a moderate expression of the HBx protein in 10-day-old animals and a lack of detection of HBx at later time points (90-day-old mice) (16).
Six-week-old female severe combined immunodeficient (SCID) mice (IFFRA-CREDO, Lyon, France) were used as the recipients of hepatocytes isolated from HBx-LacZ and LacZ transgenic mice, to minimize any risk of cell rejection.
Liver Cell Isolation, Transplantation, and Partial Hepatectomy.
Hepatocytes were isolated from 10-, 15-, and 90-day-old HBx-LacZ and LacZ mice by using collagenase (Collagenase Hepatocyte Qualified—GIBCO/BRL), as described (58). SCID mice were anesthetized with xylazine (Bayer, Leverkusen, Germany) and ketamine (Rhône Mérieux, Lyon, France) dissolved in PBS, spleens were exteriorized through a small, left-flank incision, and a syringe with a 26-gauge needle was used to inject 100 μl of cell suspension (5 × 105 to 1 × 106 viable hepatocytes) in Williams medium (GIBCO/BRL). Recipient SCID mice were held for 30 days to allow sufficient time for the proliferation and reorganization of donor hepatocytes into the liver parenchyma, before partial hepatectomy was performed. During this procedure, the left lobe and part of the median lobe of the liver were removed through a mid-abdominal incision (59). All animals were killed 8 days after partial hepatectomy.
Serum Injection After Partial Hepatectomy.
Serum was obtained from 10-, 15-, and 90-day-old HBx-LacZ mice, and 100 μl serum was injected into the spleens of SCID mice 1 day after partial hepatectomy. The animals were killed 8 days after partial hepatectomy.
Detection of Transplanted Liver Cells.
PCR analysis.
Genomic DNA was extracted from frozen liver tissues by proteinase K digestion, phenol extraction, and ethanol precipitation. LacZ genomic sequences were detected in transplanted SCID mice livers by PCR using specific LacZ primers (sense 5′-TCACACTACGTCTGAACGTCG-3′; and antisense 5′-CTGCTTCATCAGCAGGATATCC-3′). PCR was performed with 30 amplification cycles of 30 s each at the following temperatures: 94°C, 57°C, and 72°C. Murine genomic DNA was detected by amplifying the murine c-mos protooncogene as described (60).
Hepatocyte DNA staining using Hoechst fluorochrome.
After isolation, viable hepatocytes were resuspended in 2 ml of Williams medium (GIBCO/BRL), and incubated for 30 min at 37°C with 3 μl of Hoechst 33258 (10 mg/ml) (Sigma). After the inhibition of Hoechst fluorochrome with 2 ml of fetal calf serum (FCS), the cell suspensions were centrifuged three times at 50 g, resuspended in Williams medium (GIBCO/BRL) and 10% FCS, and then transplanted into recipient SCID mice by intrasplenic injection. Eight days after transplantation, liver samples were obtained by partial hepatectomy, frozen sections were cut (7 μm) with a cryostat, and Hoechst labeling was visualized under a fluorescence microscope. Three independent fields were analyzed.
In vivo detection of LacZ expression.
LacZ expression was tested on 4% paraformaldehyde-fixed, paraffin-embedded liver sections (5 μm) obtained during and 8 days after partial hepatectomy by using anti-LacZ antibodies (Rockland, Gilbertsville, PA), as described (61).
Evaluation of Liver Regeneration.
Overall liver regeneration was evaluated by weighing the mice and remaining livers after euthanasia, and by analyzing DNA synthesis by using 5-bromo-2′-deoxyuridine (BrdUrd) incorporation as described (16). BrdUrd (Sigma) was injected i.p. 24 h and 36 h after partial hepatectomy at a rate of 0.1 mg/g of body weight. The animals were killed 48 h after partial hepatectomy. Three independent fields were counted.
In Situ Detection of Apoptosis After Partial Hepatectomy.
The presence of apoptosis was sought in SCID mice transplanted with HBx-LacZ and LacZ hepatocytes. The liver cells of SCID mice undergoing apoptosis were labeled by using in situ nick end labeling of nucleosomal DNA (TUNEL), as described (16). Apoptotic hepatocytes were quantified by counting five consecutive optical fields.
RNase Protection Assay (RPA).
Total RNA was extracted from livers by using RNAble (Eurobio, Paris), following manufacturer's instructions. [α-32P]UTP-labeled antisense RNA transcripts prepared from mCK3 DNA template (Riboquant, PharMingen), using a MAXIscript in vitro transcription kit (Ambion, Austin, TX), were hybridized with 20 μg of total RNA overnight at 56°C, as per manufacturer's instructions. Bands were detected by phosphoimaging using image quant software (Molecular Dynamics). Results were calculated as a ratio of the volume of the band of interest to the mean of the volumes of the bands for the housekeeping genes encoding the large ribosomal protein L32 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Effect of HBx-Conditioned Medium on DNA Synthesis in Primary Rat Hepatocytes.
HuH7 human hepatoma cells were transfected with 20 μg of the HBx Myc-tagged-expressing plasmid, or empty vector, by using the calcium phosphate precipitation technique. Sixteen hours after transfection, the cells were washed and fresh medium was added. Conditioned medium was harvested 48 h posttransfection. Transient expression of HBx protein was confirmed by Western blot analysis (9). Primary hepatocytes were isolated from 5-week-old male Wistar rats and cultured, as described (62). After plating, conditioned media were added. In some experiments HBx- and neo-expressing HuH7 cell supernatants were first incubated either with 10 μg/ml of rabbit anti-HBx antiserum (kind gift of P. Arbuthnot, University of Witwatersrand, Parktown, South Africa), or with 2 μg/ml monoclonal anti-Myc antibody (Santa Cruz Biotechnology) for 2 h and then pulled down with protein G-Sepharose beads (Amersham Pharmacia Biotech) before being added to primary rat hepatocytes. BrdUrd was added 12 h before cell fixation in accordance with the manufacturer's instructions (Roche Molecular Biochemical). The percentage of cells undergoing DNA synthesis was estimated by counting the number of BrdUrd-labeled cells.
Statistical Analysis.
The data are presented as means ± standard deviations (SD). Statistical analysis was performed by using a nonparametric Student's t test (P < 0.05).
Results
The aim of the study was to assess the effect of HBx expression on liver cell proliferation in vivo in the context of liver regeneration after partial hepatectomy. Experiments were carried out in both HBx-LacZ and LacZ transgenic mice, as well as in SCID mice transplanted with hepatocytes isolated from these transgenic mice. We took advantage of the LacZ transgene present in transgenic mice to monitor the fate of transplanted liver cells. This LacZ sequence was also used as an internal control of the biological effects of HBx, because we could comparatively investigate hepatocytes derived from HBx-LacZ and LacZ transgenic mice.
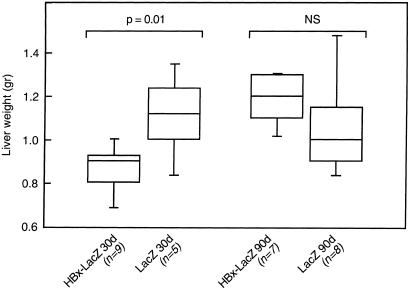
In HBx-LacZ transgenic animals, we first determined whether the expression of HBx might modify the degree of liver regeneration after partial hepatectomy. Experiments were performed at two time points (30 and 90 days after birth) in HBx-LacZ mice to evaluate the impact of moderate (30-day-old animals) or absent (90-day-old animals) HBx expression on the results. At each time point, we compared the weight of the remaining liver 8 days after partial hepatectomy in HBx-LacZ and LacZ transgenic mice. Partial hepatectomy performed in 30-day-old animals induced a 23% reduction in mouse liver weight in HBx-LacZ transgenic animals (0.86 ± 0.12 g) when compared with LacZ mice (1.11 ± 0.19 g) (P = 0.01). In contrast, no significant difference was observed in liver weight in HBx-LacZ (1.19 ± 0.12 g) and LacZ (1.06 ± 0.25 g) animals when partial hepatectomy was performed at 90 days after birth (Fig. 1). This result shows that the expression of HBx inhibited overall liver regeneration after partial hepatectomy.
Figure 1.
Inhibition of liver regeneration after partial hepatectomy in HBx-LacZ transgenic mice. Box plots of the liver weights of transgenic mice 8 days after partial hepatectomy. Partial hepatectomy was performed on HBx-LacZ or LacZ transgenic mice at days 30 or 90 after birth, as described in Materials and Methods. Each box has its ends at the quartiles, and the median of distribution is marked by a line within the box. n corresponds to the number of animals in each group.
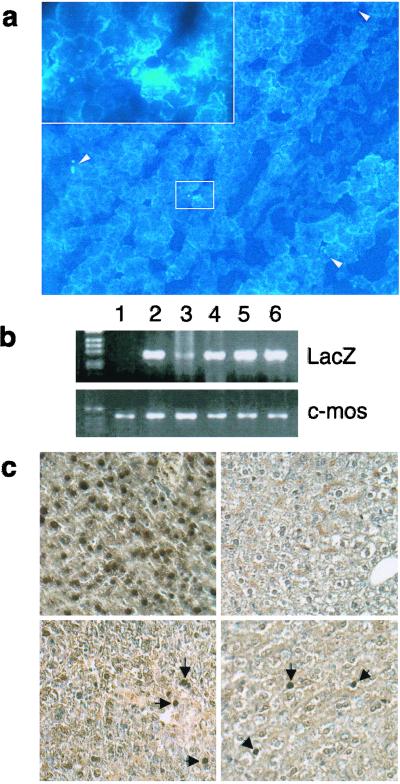
We then set up an in vivo experimental model to test for the effect on overall liver regeneration of HBx expression in a minority of liver cells. We thus performed liver cell transplantation of hepatocytes isolated from HBx-LacZ and LacZ transgenic mice, and then tested in the SCID recipient mice the extent of liver regeneration after partial hepatectomy. Two complementary approaches were used to assess liver cell transplantation. First, we visualized transplanted liver cells by using Hoechst staining. As shown in Fig. 2a, Hoechst-positive cells were detected in the liver of recipient mice 8 days after intra-splenic transplantation in the absence of partial hepatectomy. A semiquantitative estimation indicated that transplanted cells constituted less than 1/1000 in the recipient livers. Second, we took advantage of the presence of the LacZ sequence in both the HBx-LacZ and LacZ transgenic mouse liver cells used for transplantation to perform PCR analysis. As shown in Fig. 2b, LacZ sequences were indeed detected by PCR in recipient SCID liver after partial hepatectomy, thus confirming liver cell transplantation. Moreover, LacZ expression was shown in a limited number of liver cells (fewer than 1/1000) without preferential distribution in the liver sections obtained 8 days after partial hepatectomy (Fig. 2c). These results demonstrated that transplanted cells persisted upon stimulation of recipient liver regeneration and retained their transgene expression.
Figure 2.
Detection of transplanted hepatocytes in recipient liver. (a) Hoechst 33258-positive cells were detected in frozen liver sections from SCID recipient mice 8 days after intrasplenic transplantation. (Magnification, ×100.) In the upper left corner is a ×400 magnification of the selected area. (b) PCR analysis for LacZ gene or murine-specific c-mos protooncogene. Genomic DNA was extracted from the liver of SCID mouse (lane 1) and from the liver of donor HNF1-LacZ transgenic mice (lane 2). The LacZ sequence was detected in genomic DNA from transplanted SCID liver with LacZ hepatocytes (lanes 3 and 4) and HBx-LacZ hepatocytes (lanes 5 and 6). (c) In vivo detection of LacZ expression by immunohistochemical staining. (Upper Left) HBx-LacZ donor mouse liver (positive control). (Upper Right) Untransplanted SCID mouse liver (negative control). (Lower Left) SCID mouse liver transplanted with HBx-LacZ hepatocytes. (Lower Right) SCID mouse liver transplanted with LacZ hepatocytes.
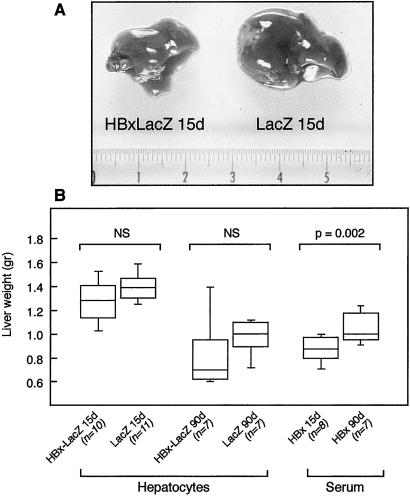
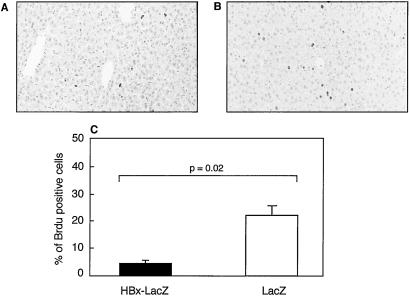
We then assessed the effects of the intrahepatic implantation of liver cells concerning the extent of liver regeneration after partial hepatectomy. Macroscopic evaluation of the liver 8 days after partial hepatectomy showed a marked reduction in liver mass in 8 of 10 recipient mice transplanted with liver cells from 10- to 15-day-old HBx-LacZ mice, compared with 2 of 11 mice transplanted with 10- to 15-day-old LacZ mouse liver cells (see representative results in Fig. 3A). In contrast, no macroscopic modification was observed when mice transplanted with 90-day-old HBx-LacZ or LacZ hepatocytes were compared. These findings, although clearly reproducible, were not confirmed by liver weight measurements (Fig. 3B). However, BrdUrd incorporation analysis performed 48 h after partial hepatectomy did confirm a marked reduction (5.5-fold) in cellular DNA synthesis after the transplantation of HBx-expressing hepatocytes isolated from 10- to 15-day-old transgenic mice (P < 0.002) (Fig. 4C). Thus, the transplantation of around 106 hepatocytes was sufficient to inhibit liver regeneration after partial hepatectomy. Standard histological examination of the liver did not reveal any obvious morphological changes (data not shown). Thus, to determine whether increased liver cell apoptosis might contribute to the inhibition of liver regeneration, we performed a terminal deoxynucleotide transferase-mediated dUTP end labeling (TUNEL) assay. There was no apparent increase in the percentage of apoptotic liver cells 1, 2, or 8 days after partial hepatectomy in mice transplanted with HBx-LacZ hepatocytes (data not shown). Therefore, the reduction in cellular DNA synthesis mostly accounted for the inhibition of liver regeneration.
Figure 3.
Inhibition of liver regeneration in SCID mice transplanted with hepatocytes or injected with serum from HBx-LacZ or LacZ transgenic mice. (A) Macroscopic evaluation of the livers of SCID mice transplanted with hepatocytes from 10- to 15-day-old HBx-LacZ transgenic mice (HBx-LacZ 15d) or 10- to 15-day-old LacZ transgenic mice (LacZ 15d) 8 days after partial hepatectomy. (B) Box plots of the liver weights of transplanted (hepatocytes) or injected (serum) SCID mice after partial hepatectomy. Transplanted hepatocytes were obtained from 10- to 15-day-old HBx-LacZ mice (HBx-LacZ 15d), or 10- to 15-day-old LacZ mice (LacZ 15d) or from 90-day-old HBx-LacZ mice (HBx-LacZ 90d) or 90-day-old LacZ mice (LacZ 90d). Serum was obtained from 10- to 15-day-old HBx-LacZ mice (HBx 15d) or 90-day-old HBx-LacZ mice (HBx 90d).
Figure 4.
Inhibition of liver regeneration by in vivo HBx expression. Immunohistochemical detection of S-phase hepatocytes 48 h after partial hepatectomy by BrdUrd incorporation in SCID mice transplanted with liver cells from 10- to 15-day-old HBx-LacZ transgenic mice (A) and SCID mice transplanted with liver cells from 10- to 15-day-old LacZ transgenic mice (B). BrdUrd-positive nuclei were stained in dark brown. (C) In situ S-phase index 48 h after partial hepatectomy in SCID mice transplanted with 10- to 15-day-old HBx-LacZ liver cells and with 10- to 15-day-old LacZ liver cells.
This inhibition of overall liver regeneration with the expression of HBx in only a small proportion of liver cells strongly suggests that HBx acted on both HBx-expressing cells and HBx-negative liver cells. To investigate this further, we tested whether the injection of serum from HBx-expressing transgenic mice might have the same effect as transplantation on liver regeneration. We therefore compared the weights of remnant livers 8 days after partial hepatectomy in SCID mice that had been injected at day 1 after partial hepatectomy with 100 μl of serum obtained from HBx-LacZ 10- to 15- and 90-day-old mice. Fig. 3B shows a 17% lowering of liver weight in mice injected with serum from 10- to 15-day-old HBx-LacZ (0.88 ± 0.11 g), compared with that seen in HBx-LacZ 90-day-old transgenic mice (1.05 ± 0.14 g) (P = 0.02). This observation was consistent with HBx triggering the secretion in serum of a factor capable of inhibiting liver regeneration. Several cytokines have been involved in liver regeneration control. We tested, therefore, whether in vivo HBx expression, both in HBx-positive transgenic mice and in mice transplanted with HBx-expressing liver cells, might modify intrahepatic cytokine RNA expression. We did not show any significant difference in TNF α and β, interleukin-6 (IL-6), IFN γ and β, or TGFβ1 and -β2 RNA expression (data not shown).
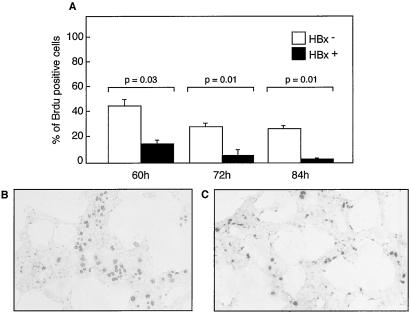
To confirm in vitro that HBx may act by a paracrine effect, we tested isolated primary rat hepatocytes on whether the in vitro ectopic expression of HBx might induce secretion into the medium of a soluble factor capable of mimicking our in vivo observation. Fig. 5 shows that the addition to primary rat hepatocytes of an HBx-expressing HuH7 cell supernatant, and not a control supernatant, induced a marked reduction in BrdUrd incorporation, 60 [3.2-fold (P = 0.003)], 72 [5.6-fold (P = 0.001)], and 84 [20-fold (P = 0.001)] h after plating.
Figure 5.
Inhibition of cell proliferation by in vitro HBx expression. (A) BrdUrd incorporation in cultured primary rat hepatocytes incubated for the indicated times with the supernatant of HuH7 cells transfected with the HBx expression vector (HBx+) or control plasmid (HBx-). (B and C) Representative results of light microscopy examination of primary rat hepatocytes incubated for 60 h with HBx- (B) or HBx+ (C) culture medium. BrdUrd was visualized by immunostaining with anti-BrdUrd antibodies.
Given these results, we tested for a direct effect of HBx on hepatocyte proliferation. Thus, we investigated the effect of HBx-expressing cell supernatant previously incubated with polyclonal anti-HBx antibodies on BrdUrd incorporation. However, we failed, in our experimental conditions, to obtain conclusive results (data not shown). Thus, as a complementary approach, we took advantage of Myc-tag present in HBx-expressing sequence and tested whether incubation of HBx-expressing cell supernatant with anti-Myc antibodies would abrogate the inhibitory effect of the supernatant on primary rat hepatocyte DNA synthesis. We failed, however, to show such abrogation (data not shown).
Overall, the results of in vivo and in vitro experiments were entirely consistent and showed a direct or indirect paracrine action of HBx. Further studies are clearly needed to dissect the fine mechanisms of such HBx-dependent effect, but the present data point to the induction, upon HBx expression, of a protein acting on adjacent liver cells.
Discussion
Taken together, our results demonstrate another mechanism for the biological effects of HBx and lead us to propose HBV as one of the few viruses implicated in human cancer that act, at least in part, through paracrine biological pathways.
In HBx-expressing mice, either transgenic or transplanted with HBx-expressing hepatocytes, we were able to demonstrate that HBx could inhibit the in vivo liver regeneration triggered by partial hepatectomy. We took advantage of the different levels of intracellular HBx expression observed in the young and older transgenic mice used for this study and showed that this effect was observed only in the context of intracellular HBx accumulation. Furthermore, these results were obtained in the context of moderate HBx expression. That HBx inhibits cell proliferation is consistent with the results of several in vitro studies (39–46, 48, 63). Thus, the in vitro ectopic expression of HBx can induce cell cycle arrest and liver cell apoptosis and, in vivo, the detection of liver cell apoptosis could be correlated, in the HBx-expressing transgenic mice utilized in our study, with HBx accumulation, as observed in young, but not in older, animals (16). In our present experimental model, the regeneration of whole recipient liver after partial hepatectomy was markedly reduced, as assessed by morphological inspection and liver weight measurements. BrdUrd incorporation and TUNEL assays showed that this reduction in liver regeneration was mostly because of a decrease in liver cell DNA synthesis, whereas we did not detect any increase in liver cell apoptosis. This result suggests that, at least under the in vivo stimulation of liver cell proliferation, the principal overall effect of moderate HBx accumulation is cell cycle arrest.
Strikingly, the results of our experiments performed to compare the transplantation of hepatocytes and the injection of serum samples from HBx-expressing transgenic mice established that HBx-dependent biological effects could not be accounted for solely by the intracellular effects of HBx, because the transplantation of about 5 × 105 HBx-expressing hepatocytes was sufficient to reduce liver regeneration. Importantly, this inhibitory effect was also achieved by injecting serum samples from HBx-expressing mice. Our in vitro experiments, demonstrating an inhibitory effect of HBx-expressing cell supernatants on primary rat hepatocyte DNA synthesis, provided additional arguments in favor of a paracrine action for HBx. Indeed, the results obtained after partial hepatectomy in HBx-expressing transgenic mice, in nontransgenic mice, and in mice either transplanted with HBx-positive hepatocytes or injected with serum from HBx-transgenic animals were entirely consistent and led us to conclude that HBx would exert an inhibitory paracrine effect on liver regeneration.
A paracrine effect of viral proteins on cell transformation and angiogenesis has been hypothesized for only a few human viruses implicated in carcinogenesis. Thus, the Kaposi sarcoma-associated herpes virus (KSHV or HHV8) acts in part through the stimulation of uninfected endothelial cells, and particularly through the secretion of vascular endothelial growth factor (VEGF) receptor 2 (64, 65), the HIV Tat (66) and HTLV-I Tax (67) proteins induce the expression and secretion of cytokines that stimulate adjacent cell proliferation and gene transactivation. It has also been suggested that HHV8 may participate in myeloma by inducing IL6 secretion (68, 69). In all these conditions, viral infection in a limited percentage of cells triggers the proliferation of adjacent cells through the secretion of either a viral protein or growth factors/cytokines. Our results provide an alternative model whereby a viral protein may in fact induce an inhibitory effect on the proliferation of uninfected cells. The nature of the inhibitory signal triggered by HBx remains unclear. Several cytokines and growth factors have been shown to regulate liver cell proliferation in a positive fashion, although only a few candidate molecules have so far been identified as inhibiting liver regeneration. Of these, we investigated the potential implication of TGFβ, given its importance to the negative regulation of liver regeneration after partial hepatectomy (53, 70), the frequent deregulation of the TGFβ-related pathway in different models of liver carcinogenesis, and the documented activation of TGFβ expression by HBx (71). However, we found no evidence of any increase in TGFβ intrahepatic mRNA expression and its secretion in serum of HBx-transgenic mice or in culture media of HBx-transfected cells (data not shown). On the other hand, it is plausible that HBx itself may be secreted; anti-HBx antibodies can be detected at all stages of HBV infection and there is also evidence for a cellular immune response to this viral protein (72). It should, however, be noted that our results were obtained in the context of only a moderate accumulation of HBx, which may rather suggest the role of an inhibitory molecule whose expression would be triggered by HBx. Also of note, we failed to abrogate the inhibitory effect of HBx-conditioned medium by using an anti-Myc antibody to deplete Myc-tagged HBx potentially present in HBx-conditioned medium (data not shown).
A large number of studies on experimental liver carcinogenesis in mice and rats have established the importance for achieving efficient tumor induction of combining the administration of a genotoxic compound with the inhibition of normal liver cell growth, thus triggering the clonal expansion of cells able to resist this inhibitory signal (73–76). Our findings lead us to propose that a viral protein implicated in human carcinogenesis, namely HBx, may in fact act by combining intracellular and paracrine effects, and inhibiting the proliferation of both infected and uninfected liver cells in the context of liver regeneration. This dual property would have a selective effect, promoting the expansion of cells resistant to the inhibitory effect of HBx. Depending on the experimental conditions, HBx has also been reported to stimulate cellular DNA synthesis and inhibit apoptosis. Moreover, we have demonstrated the biological impact of the HBx mutations observed in HCC tumor cells that abrogate the proapoptotic effects of HBx and, possibly, favor cell transformation (9). It is therefore plausible that, at later stages during the course of HCC development, modifications of HBx intracellular expression and/or HBx mutations within infected cells, as well as changes to the differentiation status of liver cells (8), may favor the expansion of HBx-expressing cells. Although these observations were made in the specific context of liver regeneration triggered by partial hepatectomy, they may represent an important mechanism of HBV-dependent liver carcinogenesis in the context of chronic hepatitis and the associated stimulation of liver cell proliferation. Finally, our results suggest that this combination of transgenic- and cell transplantation-based studies may constitute an important experimental approach in the future, for example, in the context of studies aiming to evaluate in vivo the importance of natural HBx mutants (9).
Acknowledgments
We thank Olivier Brégerie and Annie Masson for their technical support. J.G.T. was supported by a grant from the Technology and Science Foundation, Praxis, Portugal. C. Giannini was supported by a fellowship from the European Association for the Study of the Liver. This work was supported by the Institut National de la Santé et de la Recherche Médicale, and the Association pour la Recherche sur le Cancer, Ligue Nationale Contre le Cancer.
Abbreviations
- HBV, hepatitis B virus
HBx, hepatitis B virus X protein
- HCC
hepatocellular carcinoma
- WHV
woodchuck hepatitis virus
- HNF-1
hepatocyte nuclear factor 1
- SCID
severe combined immunodeficient
- TGFβ
transforming growth factor β
- TNF α
tumor necrosis factor α
- TUNEL, terminal deoxynucleotide transferase-mediated dUTP end labeling.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P. Semin Cancer Biol. 2000;10:211–231. doi: 10.1006/scbi.2000.0321. [DOI] [PubMed] [Google Scholar]
- 2.Seeger C, Mason W S. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feitelson M A. J Cell Physiol. 1999;181:188–202. doi: 10.1002/(SICI)1097-4652(199911)181:2<188::AID-JCP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Chisari F V. Am J Pathol. 2000;156:1117–1132. doi: 10.1016/s0002-9440(10)64980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F V. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 6.Minami M, Poussin K, Bréchot C, Paterlini-Bréchot P. Genomics. 1995;29:403–408. doi: 10.1006/geno.1995.9004. [DOI] [PubMed] [Google Scholar]
- 7.Chami M, Gozuacik D, Saigo K, Capiod T, Falson P, Lecoeur H, Urashima T, Beckmann J, Gougeon M L, Claret M, et al. Oncogene. 2000;19:2877–2886. doi: 10.1038/sj.onc.1203605. [DOI] [PubMed] [Google Scholar]
- 8.Andrisani O M, Barnabas S. Int J Oncol. 1999;15:373–379. doi: 10.3892/ijo.15.2.373. [DOI] [PubMed] [Google Scholar]
- 9.Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, Paterlini-Bréchot P, Bréchot C, Kremsdorf D. Cancer Res. 2001;61:7803–7810. [PubMed] [Google Scholar]
- 10.Diao J, Garces R, Richardson C D. Cytokine Growth Factor Rev. 2001;12:189–205. doi: 10.1016/s1359-6101(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 11.Schluter V, Meyer M, Hofschneider P H, Koshy R, Caselmann W H. Oncogene. 1994;9:3335–3344. [PubMed] [Google Scholar]
- 12.Soussan P, Garreau F, Zylberberg H, Ferray C, Bréchot C, Kremsdorf D. J Clin Invest. 2000;105:55–60. doi: 10.1172/JCI8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoulim F, Saputelli J, Seeger C. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim C M, Koike K, Saito I, Miyamura T, Jay G. Nature (London) 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 15.Klein N P, Schneider R J. Mol Cell Biol. 1997;17:6427–6436. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terradillos O, Billet O, Renard C A, Levy R, Molina T, Briand P, Buendia M A. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 17.Slagle B L, Lee T H, Medina D, Finegold M J, Butel J S. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 18.Sirma H, Weil R, Rosmorduc O, Urban S, Israel A, Kremsdorf D, Bréchot C. Oncogene. 1998;16:2051–2063. doi: 10.1038/sj.onc.1201737. [DOI] [PubMed] [Google Scholar]
- 19.Dandri M, Petersen J, Stockert R J, Harris T M, Rogler C E. J Virol. 1998;72:9359–9364. doi: 10.1128/jvi.72.11.9359-9364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doria M, Klein N, Lucito R, Schneider R J. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benn J, Schneider R J. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kekule A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Nature (London) 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 23.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 24.Tarn C, Lee S, Hu Y, Ashendel C, Andrisani O M. J Biol Chem. 2001;276:34671–34680. doi: 10.1074/jbc.M104105200. [DOI] [PubMed] [Google Scholar]
- 25.Weil R, Sirma H, Giannini C, Kremsdorf D, Bessia C, Dargemont C, Bréchot C, Israel A. Mol Cell Biol. 1999;19:6345–6354. doi: 10.1128/mcb.19.9.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haviv I, Vaizel D, Shaul Y. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 28.Haviv I, Shamay M, Doitsh G, Shaul Y. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qadri I, Maguire H F, Siddiqui A. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda H, Ullrich S J, Gangemi J D, Kappel C A, Ngo L, Feitelson M A, Jay G. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 31.Sitterlin D, Lee T H, Prigent S, Tiollais P, Butel J S, Transy C. J Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forgues M, Marrogi A J, Spillare E A, Wu C G, Yang Q, Yoshida M, Wang X W. J Biol Chem. 2001;276:22797–22803. doi: 10.1074/jbc.M101259200. [DOI] [PubMed] [Google Scholar]
- 34.Sun B S, Zhu X, Clayton M M, Pan J, Feitelson M A. Hepatology. 1998;27:228–239. doi: 10.1002/hep.510270135. [DOI] [PubMed] [Google Scholar]
- 35.Elmore L W, Hancock A R, Chang S F, Wang X W, Chang S, Callahan C P, Geller D A, Will H, Harris C C. Proc Natl Acad Sci USA. 1997;94:14707–14712. doi: 10.1073/pnas.94.26.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J, Duan L X, Sun B S, Feitelson M A. J Gen Virol. 2001;82:171–182. doi: 10.1099/0022-1317-82-1-171. [DOI] [PubMed] [Google Scholar]
- 37.Shih W L, Kuo M L, Chuang S E, Cheng A L, Doong S L. J Biol Chem. 2000;275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 38.Gottlob K, Fulco M, Levrero M, Graessmann A. J Biol Chem. 1998;273:33347–33353. doi: 10.1074/jbc.273.50.33347. [DOI] [PubMed] [Google Scholar]
- 39.Chirillo P, Pagano S, Natoli G, Puri P L, Burgio V L, Balsano C, Levrero M. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergametti F, Prigent S, Luber B, Benoit A, Tiollais P, Sarasin A, Transy C. Oncogene. 1999;18:2860–2871. doi: 10.1038/sj.onc.1202643. [DOI] [PubMed] [Google Scholar]
- 41.Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon M L, Tiollais P, Buendia M A. Oncogene. 1998;17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- 42.Kim H, Lee H, Yun Y. J Biol Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 43.Su F, Schneider R J. Proc Natl Acad Sci USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirma H, Giannini C, Poussin K, Paterlini-Bréchot P, Kremsdorf D, Bréchot C. Oncogene. 1999;18:4848–4859. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y C, Song K S, Yoon G, Nam M J, Ryu W S. Oncogene. 2001;20:16–23. doi: 10.1038/sj.onc.1203840. [DOI] [PubMed] [Google Scholar]
- 46.Schuster R, Gerlich W H, Schaefer S. Oncogene. 2000;19:1173–1180. doi: 10.1038/sj.onc.1203417. [DOI] [PubMed] [Google Scholar]
- 47.Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, Kurokawa K. Hepatology. 1994;19:810–819. [PubMed] [Google Scholar]
- 48.Koike K, Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Tsutsumi T, Kimura S. Cancer Lett. 1998;134:181–186. doi: 10.1016/s0304-3835(98)00252-3. [DOI] [PubMed] [Google Scholar]
- 49.Poussin K, Dienes H, Sirma H, Urban S, Beaugrand M, Franco D, Schirmacher P, Bréchot C, Paterlini-Bréchot P. Int J Cancer. 1999;80:497–505. doi: 10.1002/(sici)1097-0215(19990209)80:4<497::aid-ijc3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 50.Yu D Y, Moon H B, Son J K, Jeong S, Yu S L, Yoon H, Han Y M, Lee C S, Park J S, Lee C H, et al. J Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 51.Su Q, Schroder C H, Hofmann W J, Otto G, Pichlmayr R, Bannasch P. Hepatology. 1998;27:1109–1120. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- 52.Billet O, Grimber G, Levrero M, Seye K A, Briand P, Joulin V. J Virol. 1995;69:5912–5916. doi: 10.1128/jvi.69.9.5912-5916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fausto N. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 54.Gagandeep S, Rajvanshi P, Sokhi R P, Slehria S, Palestro C J, Bhargava K K, Gupta S. J Pathol. 2000;191:78–85. doi: 10.1002/(SICI)1096-9896(200005)191:1<78::AID-PATH587>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 55.Weglarz T C, Sandgren E P. Proc Natl Acad Sci USA. 2000;97:12595–12600. doi: 10.1073/pnas.220430497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bach I, Pontoglio M, Yaniv M. Nucleic Acids Res. 1992;20:4199–4204. doi: 10.1093/nar/20.16.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonnerot C, Vernet M, Grimber G, Briand P, Nicolas J F. Nucleic Acids Res. 1991;19:7251–7257. doi: 10.1093/nar/19.25.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guguen-Guillouzo C, Clement B, Baffet G, Beaumont C, Morel-Chany E, Glaise D, Guillouzo A. Exp Cell Res. 1983;143:47–54. doi: 10.1016/0014-4827(83)90107-6. [DOI] [PubMed] [Google Scholar]
- 59.Higgins G, Anderson R. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 60.Mercer D F, Schiller D E, Elliott J F, Douglas D N, Hao C, Rinfret A, Addison W R, Fischer K P, Churchill T A, Lakey J R, et al. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 61.Guidotti J E, Mallet V O, Parlier D, Mitchell C, Fabre M, Jaffray P, Lambert M, Kahn A, Gilgenkrantz H. Hepatology. 2001;33:10–15. doi: 10.1053/jhep.2001.20678. [DOI] [PubMed] [Google Scholar]
- 62.McIntyre M, Desdouets C, Senamaud-Beaufort C, Laurent-Winter C, Lamas E, Bréchot C. Oncogene. 1999;18:4577–4585. doi: 10.1038/sj.onc.1202815. [DOI] [PubMed] [Google Scholar]
- 63.Takada S, Shirakata Y, Kaneniwa N, Koike K. Oncogene. 1999;18:6965–6973. doi: 10.1038/sj.onc.1203188. [DOI] [PubMed] [Google Scholar]
- 64.Cesarman E, Mesri E A, Gershengorn M C. J Exp Med. 2000;191:417–422. doi: 10.1084/jem.191.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A, Gerhengorn M C. Nature (London) 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 66.Verhoef K, Klein A, Berkhout B. Virology. 1996;225:316–327. doi: 10.1006/viro.1996.0606. [DOI] [PubMed] [Google Scholar]
- 67.Siekevitz M, Feinberg M B, Holbrook N, Wong-Staal F, Greene W C. Proc Natl Acad Sci USA. 1987;84:5389–5393. doi: 10.1073/pnas.84.15.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szekely L, Klein G. Trends Microbiol. 1997;5:424–426. doi: 10.1016/S0966-842X(97)01152-9. [DOI] [PubMed] [Google Scholar]
- 69.Brousset P, Cesarman E, Meggetto F, Lamant L, Delsol G. Hum Pathol. 2001;32:95–100. doi: 10.1053/hupa.2001.21131. [DOI] [PubMed] [Google Scholar]
- 70.Braun L, Mead J E, Panzica M, Mikumo R, Bell G I, Fausto N. Proc Natl Acad Sci USA. 1988;85:1539–1543. doi: 10.1073/pnas.85.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee D K, Park S H, Yi Y, Choi S G, Lee C, Parks W T, Cho H, de Caestecker M P, Shaul Y, Roberts A B, Kim S J. Genes Dev. 2001;15:455–466. doi: 10.1101/gad.856201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stemler M, Weimer T, Tu Z X, Wan D F, Levrero M, Jung C, Pape G R, Will H. J Virol. 1990;64:2802–2809. doi: 10.1128/jvi.64.6.2802-2809.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laconi E. Am J Pathol. 2000;156:389–392. doi: 10.1016/S0002-9440(10)64741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulte-Hermann R. In: Toxic Injury of the Liver. Farber E, Fisher M M, editors. New York: Dekker; 1979. pp. 385–444. [Google Scholar]
- 75.Gordon G J, Coleman W B, Hixson D C, Grisham J W. Am J Pathol. 2000;156:607–619. doi: 10.1016/S0002-9440(10)64765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farber E, Sarma D S. Lab Invest. 1987;56:4–22. [PubMed] [Google Scholar]