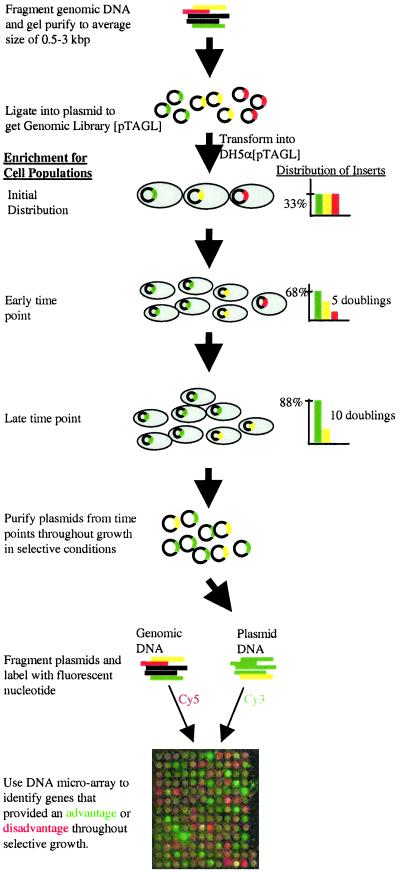
Figure 1.
PGTM. E. coli MG1655 genomic DNA was fragmented, size selected, repaired, and ligated into TOPO-TA cloning vector. E. coli DH5α were transformed with the ligation reaction, successful transformants were harvested, and their plasmids were purified. This library was used as the starting pool for all subsequent transformations. To perform the dynamic screen, chemically competent DH5α were transformed with the library and grown immediately in LB + Ampicillin (LBA) until an OD600 of 0.1–0.2 (≈5 h), at which point they were inoculated (at 1% vol/vol) into selective (0.1–2.5% vol/vol Pine-Sol) and nonselective LBA. Plasmid samples were obtained throughout growth, fragmented by sonication, labeled with Cy3, and mixed with genomic E. coli DNA labeled with Cy5. The mixture was hybridized to an E. coli DNA microarray containing 1,160 gene probes. Differences among Cy3 intensities reflect the enrichment for specific transformant subpopulations out of the heterogeneous transformant library. Transformants that contain inserts that provide a growth advantage (green) will become the majority members of the total plasmid population at the expense of transformants that contain inserts that either do not provide a growth advantage (yellow) or that provide a growth disadvantage (red). In the example bar charts, the doubling times corresponding to each transformant type were 45 min (green), 60 min (yellow), and 85 min (red). Starting from an equal distribution of transformants (33% each), transformants with green inserts will make-up close to 90% of the total cell populations after 10 doublings, whereas those with red inserts will be diluted to ≈1% of the total number of cells.