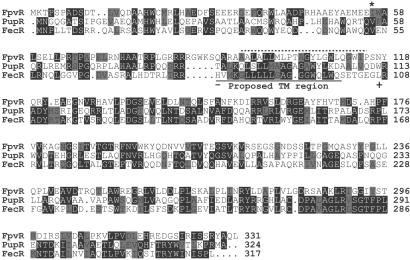
Figure 4.
Alignment of the sequences of FpvR, PupR, and FecR. The sequence of FpvR (PA2388 in the P. aeruginosa PAO1 genomic sequence) was aligned with those of PupR and FecR. Positions where identical or similar residues are present in at least two of the sequences are highlighted with identical residues shaded black and similar residues (A and G; D and E; F, W and Y; I, L, M, and V; N and Q; S and T; R and K) shaded gray. FpvR has 27.4% identity (49.0% similarity) with PupR and 35.1% identity (43.0% similarity) with FecR. Sites where fusion of β-lactamase with the N-terminal portion of FecR resulted in sensitivity (−) (cytoplasmic or intramembrane) or resistance (+) (periplasmic) to ampicillin (43) are indicated, and the predicted membrane-spanning region of FecR is underlined. A broken line indicates the region of FpvR that is predicted with TMHMM and TMPRED to constitute a transmembrane helix. The asterisk (*) corresponds to the site where a mutation was introduced into the gene encoding FpvR (see text).