FIG. 1.
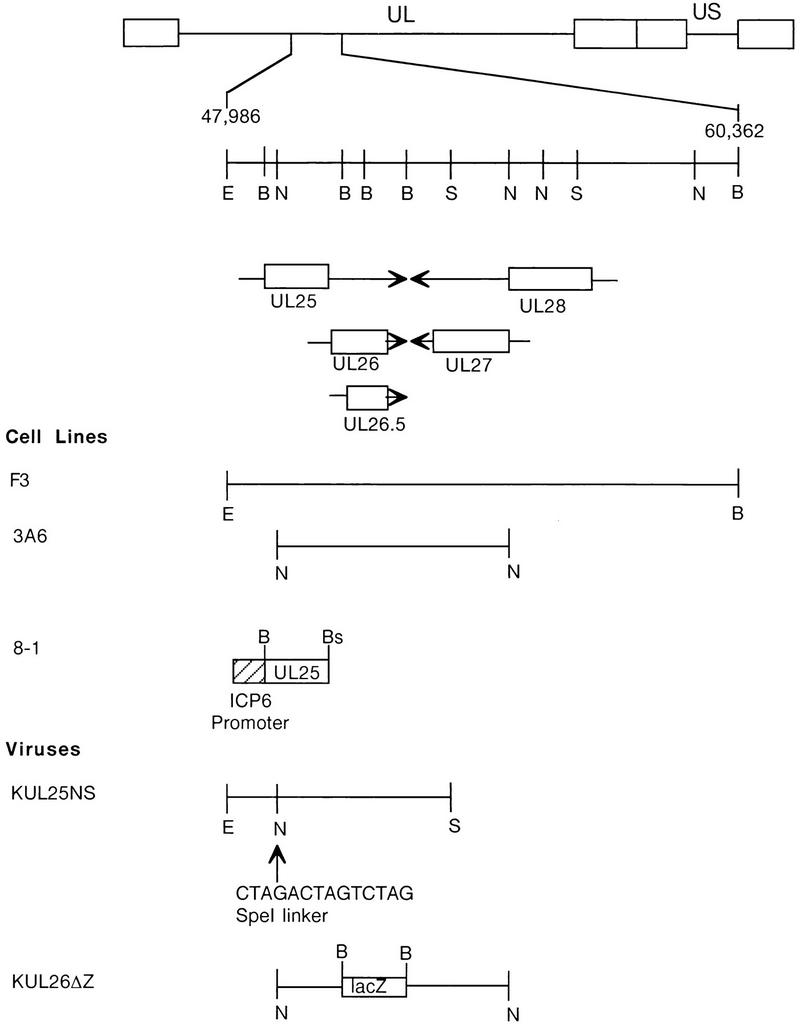
Regions of the HSV-1 genome that were used to isolate transformed cells and recombinant viruses. The HSV-1 genome is shown at the top; UL and US refer to the long and short unique region sequences. On the next line, the EcoRI-to-BamHI region located between nucleotides 47986 and 60362 of the HSV-1 genome is expanded. The locations and directions of transcription of the UL25, UL26, UL26.5, UL27, and UL28 genes are indicated in the next three lines, with the boxed regions representing the open reading frames for these genes. (Cell lines) Regions of the HSV-1 genome contained in recombinant plasmids (see Materials and Methods) used to isolate transformed Vero cell lines F3 (pKEF-B5), 3A6 (pKEF-NotI), and 8-1 (pAPV-UL25). (Viruses) Plasmid pKUL25NS, which contains an SpeI oligomer inserted within the UL25 open reading frame encoding termination codons in all three reading frames, was used to isolate the UL25 mutant KUL25NS. KUL26ΔZ is a UL26 mutant in which a lacZ cassette was inserted in place of UL26 codons 41 through 593 (19). Restriction endonuclease sites: E, EcoRI; B, BamHI; N, NotI; S, SpeI; Bs, BsrI.
