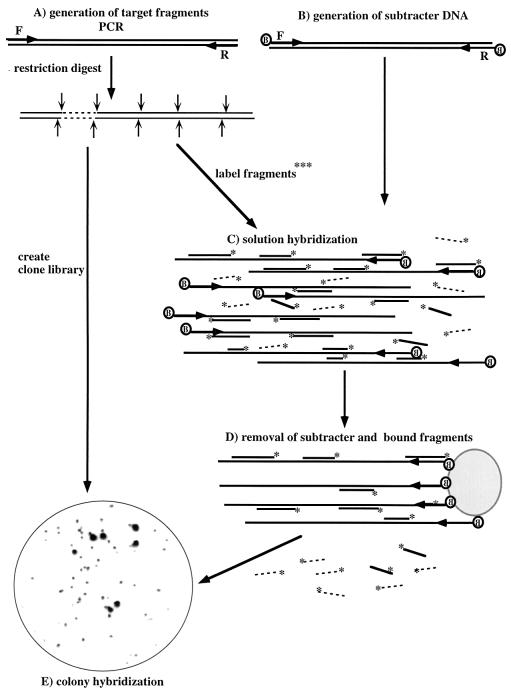
FIG. 1.
Subtractive hybridization strategy used to obtain habitat-based probes. A, Generation of restriction endonuclease-generated subfragments from PCR-amplified target DNA from a 16S rDNA clone or bacterial strain, creation of a clone library of target subfragments, and radioactive labeling of subfragments for hybridization; B, generation of biotinylated subtracter DNA by PCR amplification of 16S rDNA from the total DNA extracted from the environmental sample by using two 5′-biotinylated primers; C, solution hybridization of radioactively labeled target fragments and biotinylated subtracter DNA; D, removal of subtracter-target hybrids by binding to streptavidin-coated beads; E, use of enriched target fragments for screening of the clone library of target fragments by colony hybridization. In the example shown a clone library of subfragments of Acidovorax-like sequence SW4 was screened with subfragments enriched by two hybridizations with subtracter DNA consisting of 16S rDNA amplified from DNA extracted from Spittelwasser sediment. F and R indicate forward and reverse primers, respectively; B indicates biotin; and ∗ indicates 32P end label. Radioactive target fragments removed by subtractive hybridization are shown as solid lines, whereas those which tend not to be removed by subtractive hybridization and which are thus specific for the target are shown as dashed lines.