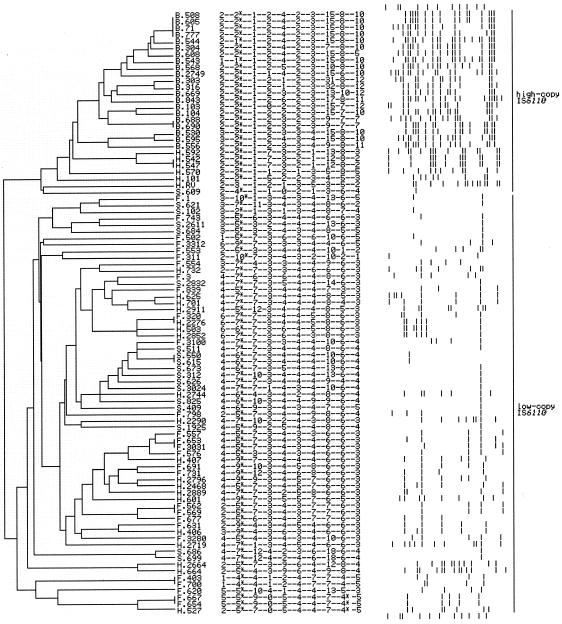
FIG. 3.
Dendrogram of genetic relationships of 92 isolates of M. tuberculosis (denoted by numbers) based on the 10 VNTR loci. The consensus tree is built using the unweighted pair group method with arithmetic averages algorithm in the PAUP program. The isolates are classified into two separate lineages: high IS6110 copy number (more than eight IS6110 copies, except for the single-banded isolate S.609 and the eight-banded isolate H.101) and low IS6110 copy number (one to eight IS6110 copies, except for the three heterogeneous isolates H.664, H.2664, and H.2719, which have more than eight IS6110 copies). Letters B, F, H, and S denote the different IS6110 types: Beijing, few-banded, heterogeneous, and single-banded isolates, respectively. The VNTR profiles are shown in the 10-digit column, representing the number of copies of the repeat units in VNTR0569, VNTR0580, VNTR0595, VNTR0960, VNTR1955, VNTR2461, VNTR3690, VNTR3820, VNTR4052, and VNTR4120, respectively. Asterisks at VNTR0580 and VNTR4052 indicate partial deletions in the VNTR sequences. The IS6110 RFLP profiles are shown, with high to low molecular weight oriented from left to right. The top and bottom rows represent the IS6110 RFLP of the Mt14323 strain.