Abstract
From December 2004 to March 2005, 27 Klebsiella pneumoniae clinical isolates that were positive by the imipenem-EDTA double-disk synergy test and that exhibited a single macrorestriction pattern were recovered in two distinct Greek hospitals. The isolates carried a transferable blaVIM-1 metallo-β-lactamase gene in a class 1 integron. Reverse transcriptase PCR showed that the gene was similarly expressed in low- and high-level carbapenem-resistant isolates, indicating the existence of additional resistance mechanisms. The clonal spread of VIM-1-producing K. pneumoniae strains in distinct regions where up to now blaVIM-2 and blaVIM-4 alleles were common is worrisome.
The acquired metallo-β-lactamases (MBLs) comprise a group of enzymes with a broad hydrolytic spectrum that includes carbapenems. Four groups of MBLs have been described up to now, namely, IMP, VIM, SPM, and GIM, with the IMP and the VIM types being prevalent and reported from various regions worldwide (3, 11). MBLs of the VIM type are more common among nonfermenting gram-negative bacteria, mainly in the Far East and Southern Europe (4). However, during the last few years, studies have reported on the dissemination of VIM-type MBLs in members of the family Enterobacteriaceae (2, 5), suggesting the ongoing spread of these resistance determinants among pathogens with higher infectivities.
Preliminary susceptibility data in our tertiary-care hospitals (University Hospital of Larissa in central Greece and Hippokration University Hospital, Thessaloniki, northern Greece) indicated that from November 2004, several infections were due to carbapenem-resistant or -intermediate Klebsiella pneumoniae strains that were positive by the imipenem-EDTA double-disk synergy test (DDST). The similar antimicrobial susceptibility patterns of these isolates prompted an investigation to determine whether the limited spread of a single strain had occurred and also to study the carbapenem resistance mechanisms.
The study included 27 nonrepetitive K. pneumoniae isolates exhibiting reduced susceptibility or resistance to carbapenems (imipenem and meropenem MICs ≥ 1 mg/liter) that were consecutively isolated between December 2004 and March 2005 at the University Hospital of Larissa and the Hippokration University Hospital. Carbapenem-nonsusceptible K. pneumoniae strains had not been isolated at these institutions up to November 2004, and these isolates were all the carbapenem-nonsusceptible K. pneumoniae isolates recovered during the study period. Seventeen isolates were recovered at the University Hospital of Larissa (16 of them were recovered from 12 patients hospitalized in an intensive care unit and two medical wards, and 1 isolate was recovered from an outpatient who had had a recent hospitalization in a private hospital in northern Greece). The remaining 10 isolates were recovered from clinical samples of separate patients hospitalized in five medical or surgical wards at Hippokration University Hospital. These two hospitals are among the largest in the country, with about 1,500 beds in total and several critical care units.
The isolates were identified to the species level by using the API 20E system (bioMérieux, Marcy l'Etoile, France). The susceptibilities of the isolates to a range of antimicrobials (imipenem, meropenem, aztreonam, cefoxitin, ceftazidime, cefepime, amikacin, netilmicin, tobramycin, gentamicin, piperacillin in combination with tazobactam, and ciprofloxacin) were determined by the disk diffusion method (7). The MICs of the strains for aztreonam, ceftazidime, piperacillin-tazobactam, cefoxitin, imipenem without and with EDTA (Etest MBL), and meropenem were determined by Etest (AB Biodisk, Solna, Sweden). Tests for possible metallo-β-lactamase production were also performed by using the imipenem-EDTA DDST (8) with the disks set 20 mm apart.
The strains were tested for the possible carriage of IMP and VIM carbapenemase genes by PCR with one and two sets of consensus primers, respectively (9, 11, 12). Primers that amplify the whole blaVIM gene were also used (14). Additionally, PCR for the detection of the blaTEM, blaCTX-M, and blaSHV genes was performed with all isolates (13) to check for the possible presence of extended-spectrum β-lactamases encoding genes commonly found in our regions.
Nucleotide sequencing of both strands of the PCR products derived with primers that amplify the whole blaVIM gene (14) was performed with an ABI Prism 377 DNA sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Mating experiments were performed by using the susceptible recipient strain Escherichia coli 26R793 (lac negative, rifampin resistant) (3). Transconjugants were selected in McConkey agar plates containing 100 μg/ml rifampin and imipenem at concentrations ranging from 0.5 to 2 μg/ml. blaVIM-bearing transconjugants were analyzed for plasmids by an alkaline lysis procedure (3). The plasmid DNA band was extracted from the agarose gel with a QIAquick gel extraction kit (QIAGEN GmbH, Hilden, Germany) and was used as the template DNA in a PCR for the detection of blaVIM. The expression of the blaVIM gene in these isolates was tested for repeatedly by reverse transcriptase PCR (RT-PCR)-specific amplification with a DNase-treated RNA extract obtained with an RNeasy mini kit (QIAGEN) and 16S rRNA as a control. The same RNA extract was used in all reactions for both 16S rRNA and blaVIM, which were run under the same conditions.
Pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA of the 27 VIM-producing K. pneumoniae strains was performed with a CHEF-DRIII system (Bio-Rad, Hemel Hempstead, United Kingdom), as described elsewhere (12). The banding patterns of the strains were compared visually.
Of the 27 isolates evaluated in the study, 10 were recovered from blood, 6 were recovered from urine, 2 were recovered from infected surgical wounds, 2 were recovered from bronchial secretions, 2 were recovered from sputum, 2 were recovered from cerebrospinal fluid, and 1 was recovered from peritoneal fluid. The MICs for imipenem of all but three isolates ranged from 1 to 16 mg/liter, and those for meropenem ranged from 1 to 4 mg/liter, while in the remaining three isolates the MICs were >32 mg/liter for both carbapenems. All isolates exhibited synergy with EDTA in the Etest MBL, which was not apparent in seven isolates with imipenem MICs of <4 mg/liter, where the interpretation of MBL production was difficult. Twenty-one isolates retained full susceptibility to aztreonam, and all were resistant to the remaining β-lactams tested. The isolates exhibited resistance to most non-β-lactam antimicrobials tested, with a few of them being susceptible to aminoglycosides or tetracycline (data not shown).
The PCR for the blaVIM gene was repeatedly positive with the two sets of consensus primers (9, 12) and negative for blaIMP in all isolates. When primers for the whole blaVIM gene were used (14), a product was amplified from all blaVIM-positive isolates. PCR for the blaSHV and blaCTX-M genes was negative for all isolates, while the blaTEM PCR was negative for all but five isolates. The latter isolates also exhibited high-level resistance to aztreonam (MICs, 48 to 128 mg/liter).
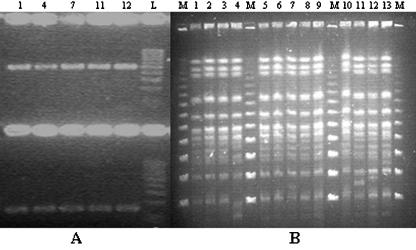
It is of interest that all isolates from both regions belonged to a single clone. Twenty-four of them were indistinguishable by PFGE, and three differed by one band (isolates 4, 11, and 13; Fig. 1B). The pattern of our unique PFGE type apparently differed from the patterns of the previously reported VIM-1-producing K. pneumoniae clones from southern Greece (2). Although our isolates were not compared in parallel with those described by Giakkoupi et al. (2), those isolates were also produced after XbaI macrorestriction.
FIG. 1.
(A) RT-PCR for blaVIM gene in the five Klebsiella pneumoniae isolates listed in Table 1 and shown in panel B; upper part of the gel, PCR products specific for blaVIM; lower part of the gel, PCR products specific for 16S rRNA. Lane L, 100-bp ladder. (B) PFGE of 13 isolates evaluated in the study after XbaI macrorestriction. Lanes M, molecular mass markers (48.5 kb).
Conjugational transfer of carbapenem resistance was tested with five isolates randomly selected from among the blaVIM producers detected in the study (Table 1). Carbapenem resistance was found to be transferable in four of the five isolates, along with resistance to aminoglycosides and other antimicrobials, with the transfer frequencies ranging from 10−2 to 10−5 per recipient cell. An apparently identical single ca. 80-kb plasmid was visualized in all transconjugant strains and also in the clinical isolate that did not transfer the resistance phenotype. The PCR for the detection of blaVIM by using as the template the gel-extracted plasmid DNA band was positive for all transconjugant strains, suggesting that the gene resided in this transferable plasmid. When both strands of the whole blaVIM gene amplicons from these isolates were sequenced, a blaVIM-1 sequence identical to that originally reported (3) was identified. The blaVIM gene was found by RT-PCR to be expressed at a similar level in all five isolates tested, which had carbapenem MICs that ranged from 2 to >32 mg/liter (Table 1; Fig. 1A).
TABLE 1.
Dates and clinical sources of isolation and resistance phenotypes of five blaVIM-1-carrying Klebsiella pneumoniae isolates and their respective transconjugantsc
| Isolateb | Clinical sample | Region | Date of isolation (mo/day/yr) | E-test MICs (μg/ml) of β-lactam antibiotics
|
Resistance to non-β-lactam antibiotics | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IPM | AZT | CAZ | TZP | FOX | |||||
| 1 | Blood | Larissa | 12/19/04 | 2 | 8 | 0.125 | >256 | 64 | >256 | CIP, TOB, SXT |
| Tcs-1a | 0.38 | 1 | 0.064 | >256 | 8 | >256 | TOB, SXT | |||
| 4 | Urine | Larissa | 12/20/04 | 4 | 8 | 48 | >256 | >256 | >256 | GEN, CIP, TOB, NET, SXT |
| Tcs-2 | 2 | 0.5 | 0.25 | 32 | >256 | >256 | GEN, TOB, NET, SXT | |||
| 7 | Blood | Larissa | 12/20/04 | >32 | >32 | 0.25 | >256 | >256 | >256 | CIP, TOB, NET, TET, SXT |
| Tcs-3 | 0.38 | 16 | 0.064 | 64 | 32 | >256 | TOB, NET, TET, SXT | |||
| 11 | Pus | Thessaloniki | 1/14/05 | 4 | 6 | 0.94 | >256 | >256 | >256 | AMK, CIP, TOB, NET, TET, SXT |
| Tcs-4 | 0.50 | 0.75 | 0.064 | >256 | 128 | >256 | AMK, TOB, NET, TET, SXT | |||
| 12 | Bronchial fluid | Larissa | 1/29/05 | 4 | 8 | 0.25 | >256 | >256 | >256 | AMK, CIP, TOB, NET, TET, SXT |
Tcs, transconjugant strain.
The carbapenem resistance of isolate 5 was nontransferable.
Abbreviations: MEM, meropenem; IPM, imipenem; AZT, aztreonam; CAZ, ceftazidime; TZP, piperacillin-tazobactam; FOX, cefoxitin; CIP, ciprofloxacin; TOB, tobramycin; SXT, trimethoprim-sulfamethoxazole; GEN, gentamicin; NET, netilmicin; TET, tetracycline; AMK, amikacin.
In one isolate (isolate 1) of the study (Table 1; Fig. 1A), PCR mapping with primers for the 5′ conserved segment of class 1 integrons (5′ CS) and for the blaVIM, aacA, dhfrI, aadA, qac, and sul genes revealed the carriage of the metallo-β-lactamase gene in a class 1 integron of the same structure as that described previously in Greece (2). Specifically, the variable region included the 5′ to 3′ cassettes containing blaVIM, aacA, and dhfrI genes. When the overlapping PCR amplicons were sequenced, it was shown that the class 1 integron contained the intI1 gene with a strong P1 promoter, followed directly by an inactivated (without a GGG insertion) P2 promoter and an attI1 site. This was followed by the blaVIM-1 gene cassette with its 59-base element that was identical to the one described previously from Italy (3). Downstream of the blaVIM-1 gene cassette was an aacA7 cassette with its 59-base element, a dhfrI cassette, and an aadA1 cassette prior to the conserved qacEΔ1 and sul1 elements.
VIM-type MBL-producing K. pneumoniae strains have increasingly been reported in Europe during the last few years (2, 5). These reports usually included sporadic cases of one or a few isolates from each hospital, while in the case of different hospitals from the same region, the strains detected were mainly unrelated (2). The present study reports on the spread of a single clone of VIM-1-producing K. pneumoniae that caused relatively large outbreaks in two geographically distinct tertiary-care hospitals during a period of a few months. It should be noted that in central and northern Greece, pseudomonads carrying the blaVIM-2 and blaVIM-4 genes are commonly isolated (6, 10, 12). The emergence also of blaVIM-1 gene in these regions indicates the wide circulation of MBL-encoding genes and poses challenges for the treatment of hospital infections due to gram-negative bacteria. All isolates tested harbored a transferable plasmid of a size similar to that carried the blaVIM-1 gene as well as traits of resistance to aminoglycosides and other antibiotics, suggesting a potential for the spread of these resistance determinants.
The apparently different macrorestriction patterns of the present clone compared with those reported previously from southern Greece (2) indicate that the clone from central and northern Greece arose independently. The single clonal outbreaks described in this study may still be restricted if urgent infection control measures are applied, prior to their evolution to polyclonal endemicity that would be very difficult to contain.
Most carbapenemase-producing K. pneumoniae isolates were susceptible to carbapenems, supporting previous findings that carbapenems maintain some clinical efficacy against MBL-positive enterobacteria, with MICs remaining below the susceptibility breakpoint (2, 5). However, a significant increase in carbapenem MICs with higher inoculum sizes has been observed previously (5), suggesting that a clinical failure of carbapenem therapy might not be unexpected. A few isolates from both regions exhibited high-level carbapenem resistance. Since the expression of the blaVIM gene was found by RT-PCR to be similar in all isolates tested, a contribution of other unrelated mechanisms, such as mutations that account for porin deficiency and impaired permeability (1), cannot be excluded. The substantially lower imipenem MICs of the transconjugants compared with those of the clinical isolates support this hypothesis.
The emergence of carbapenemases in K. pneumoniae represents a relevant clinical problem, as this pathogen may act as a reservoir for a variety of resistance plasmids. Our experience has shown that MBL detection in isolates with imipenem MICs <4 mg/liter might be difficult by Etest MBL, while DDST allowed the correct identification of all blaVIM-1-positive isolates. Therefore, screening tests with imipenem and EDTA disks might be applied in our hospital laboratories to allow the prompt detection of such highly infective multiresistant nosocomial pathogens.
REFERENCES
- 1.Crowley, B., V. J. Benedí, and A. Domenech-Sanchez. 2002. Expression of SHV-2 β-lactamase and of reduced amounts of OmpK36 porin in Klebsiella pneumoniae results in increased resistance to cephalosporins and carbapenems. Antimicrob. Agents Chemother. 46:3679-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giakkoupi, P., A. Xanthaki, M. Kanellopoulou, A. Vlahaki, V. Miriagou, S. Kontou, E. Papafraggas, H. Malamou-Lada, L. S. Tzouvelekis, N. J. Legakis, and A. C. Vatopoulos. 2003. VIM-1 metallo-β-lactamase-producing Klebsiella pneumoniae strains in Greek hospitals. J. Clin. Microbiol. 41:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livermore, D. M. 2002. The impact of carbapenemases on antimicrobial development and therapy. Curr. Opin. Investig. Drugs 3:218-224. [PubMed] [Google Scholar]
- 5.Luzzaro, F., J. D. Docquier, C. Colinon, A. Endimiani, G. Lombardi, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2004. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-β-lactamase encoded by a conjugative plasmid. Antimicrob. Agents Chemother. 48:648-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavroidi, A., A. Tsakris, E. Tzelepi, S. Pournaras, V. Loukova, and L. S. Tzouvelekis. 2000. Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46:1041-1043. [DOI] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing: thirteenth international supplement M100-S13, Table 2D. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 8.Oh, E. J., S. Lee, Y. J. Park, J. J. Park, K. Park, S. I. Kim, M. W. Kang, and B. K. Kim. 2003. Prevalence of metallo-β-lactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-β-lactamase. J. Microbiol. Methods 54:411-418. [DOI] [PubMed] [Google Scholar]
- 9.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51:1409-1414. [DOI] [PubMed] [Google Scholar]
- 11.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsakris, A., S. Pournaras, N. Woodford, M.-F. I. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzelepi, E., C. Magana, E. Platsouka, D. Sofianou, O. Paniara, N. J. Legakis, A. C. Vatopoulos, and L. S. Tzouvelekis. 2003. Extended-spectrum β-lactamase types in Klebsiella pneumoniae and Escherichia coli in two Greek hospitals. Int. J. Antimicrob. Agents 21:285-288. [DOI] [PubMed] [Google Scholar]
- 14.Yan, J.-J., P.-R. Hsueh, W.-C. Ko, K.-T. Luh, S.-H. Tsai, H.-M. Wu, and J.-J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]