Fig 2.
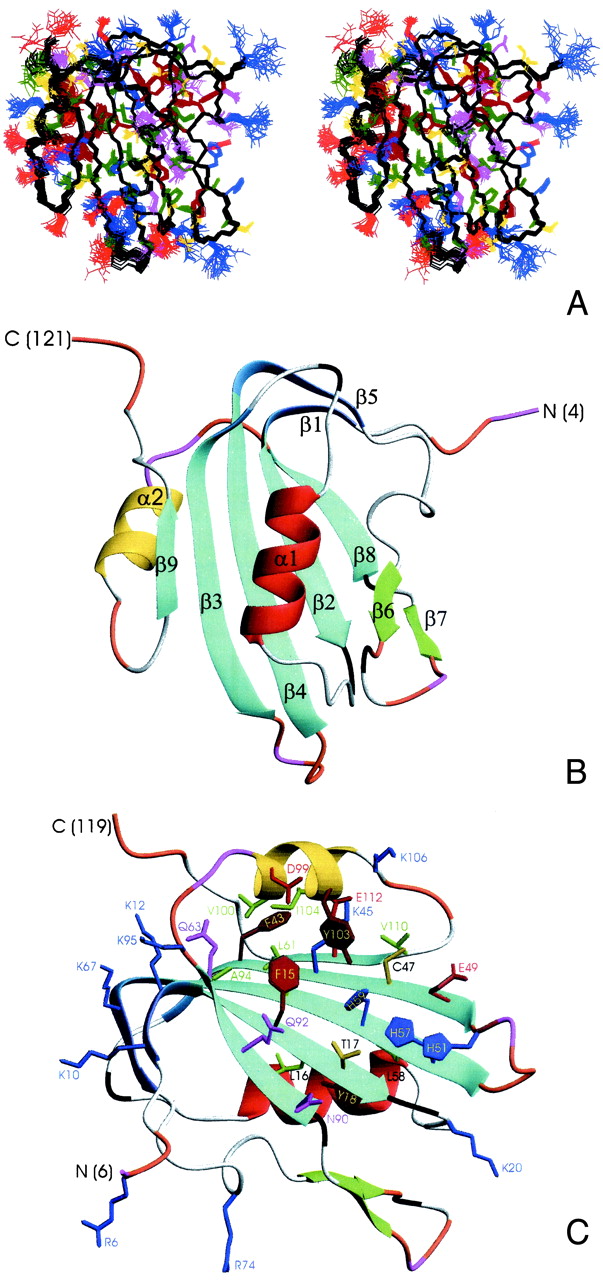
Three-dimensional structure of TYLCV Rep4–121. (A) Stereoview displaying best-fit superposition of the final ensemble (residues 6 to 119) of 30 conformers with the lowest dyana (21) target function (PDB ID ). The protein backbone (N, Cα, CO) is shown in black, and the side chains are colored according to residue type (YFW: brown; D,E: red; K,R,H: blue; A,V,L,I,P: green; T,S,C: yellow; N,Q: magenta). The coordinate precision for the protein backbone heavy atoms is 0.48 Å. (B and C) Ribbon representations of the TYLCV Rep4–121 regularized mean structure (PDB ID ). The central 5-stranded β-sheet is shown in blue, the small extension sheet in dark blue, the helix covering the β-sheet in red, the small 2-stranded sheet in green and loops in gray. The helix carrying the catalytic tyrosine is colored yellow. The strands and helices are numbered and the N and C termini labeled. Loop residues exhibiting substantial flexibility (low 15N heteronuclear NOE) or nondetected NH resonances are colored in orange and magenta, respectively. In C, selected amino acid side chains are displayed as well. They either belong to the conserved sequence motifs or occupy equivalent positions to those implicated in ss- or dsDNA/RNA binding of structurally related proteins (see Fig. 4).
