Summary
Here, we present a protocol for noninvasive, spatiotemporal visualization of hematopoietic reconstitution in live mice using Akaluc-based bioluminescence imaging (AkaBLI). We describe steps for Akaluc gene transduction into murine hematopoietic stem cells (HSCs), ex vivo expansion of hematopoietic stem and progenitor cells, and longitudinal imaging post transplantation. Representative results illustrate the spatiotemporal pattern of hematopoietic recovery in the spleen and bone marrow. This protocol enables quantitative in vivo tracking of hematopoietic reconstitution dynamics at whole-body resolution following lethal irradiation.
For complete details on the use and execution of this protocol, please refer to Yogo et al.1
Subject areas: Cell Biology, Cell culture, Stem Cells, Cell Differentiation
Graphical abstract

Highlights
-
•
Efficient lentiviral transduction of Akaluc into murine hematopoietic stem cells
-
•
Ex vivo expansion of HSCs in serum-free medium for downstream imaging studies
-
•
Intravenous transplantation of Akaluc+ HSCs into lethally irradiated recipient mice
-
•
Longitudinal in vivo imaging of hematopoietic reconstitution using Akaluc bioluminescence
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol for noninvasive, spatiotemporal visualization of hematopoietic reconstitution in live mice using Akaluc-based bioluminescence imaging (AkaBLI). We describe steps for Akaluc gene transduction into murine hematopoietic stem cells (HSCs), ex vivo expansion of hematopoietic stem and progenitor cells, and longitudinal imaging post transplantation. Representative results illustrate the spatiotemporal pattern of hematopoietic recovery in the spleen and bone marrow. This protocol enables quantitative in vivo tracking of hematopoietic reconstitution dynamics at whole-body resolution following lethal irradiation.
Before you begin
Tracking hematopoietic stem cell (HSC) behavior after transplantation is critical for understanding hematopoietic reconstitution and evaluating therapeutic efficacy. Traditional methods rely on snapshot analyses, which fail to capture dynamic changes over time. This protocol leverages the Akaluc-based bioluminescence imaging (AkaBLI) system2 to enable longitudinal, noninvasive visualization of hematopoietic reconstitution in vivo.1 By introducing the Akaluc gene into HSCs via lentiviral transduction and performing time-course in vivo imaging, researchers can quantify hematopoietic activity in organs such as the spleen and bone marrow. This approach allows comparative analysis of distinct progenitor populations and provides insight into spatiotemporal hematopoietic patterns, including the multiphasic dynamics of transplanted HSCs.
Innovation
In previous studies, analyses of hematopoietic reconstitution after transplantation have primarily relied on techniques such as immunostaining, flow cytometry, and RNA sequencing. These methods require the sacrifice of animals to obtain tissues, thereby limiting analyses to specific time points and organs, and precluding longitudinal assessment within the same individual.
This protocol offers a technical advance that enables noninvasive, spatiotemporal visualization of hematopoietic reconstitution in live mice. By integrating lentiviral gene delivery, optimized ex vivo culture of HSCs, and AkaBLI, this approach allows sensitive, whole-body tracking of hematopoietic activity over time without the need to sacrifice the animal.
Unlike conventional snapshot-based methods, this system captures dynamic aspects of hematopoietic reconstitution, including transient signal fluctuations and multiphasic recovery patterns that are often overlooked. Moreover, it enables the detection of hematopoietic activity throughout the body, rather than being restricted to predefined organs, allowing for the discovery of previously unrecognized hematopoietic sites.
Taken together, this protocol provides a robust and quantitative platform for in vivo analysis of hematopoietic stem cell behavior, assessment of functional heterogeneity, and evaluation of genetic or pharmacologic interventions. Importantly, it enables longitudinal analysis across time, enhancing both reproducibility and biological insight.
Institutional permissions
All animal studies were conducted in accordance with institutional protocols and were approved by the Animal Care and Use Committee of the Institute of Medical Science at the University of Tokyo.
Preparation of Akaluc lentiviral supernatant
Timing: 5 days
The Akaluc lentiviral vector is designed to express both mNeonGreen (mNG) and Akaluc under the control of the human UBC promoter. These two genes are linked via a self-cleaving P2A peptide, enabling bicistronic expression (Figure 1A). The procedure for generating the Akaluc lentiviral vector is described below.
-
1.DAY 0 - Day before Transfection.
-
a.Split 293T cells one day before transfection into 10 cm dish (5.0 × 106 cells per dish in 5 mL of medium x 10) using DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin-L-Glutamine (PSG).
-
b.Incubate the cells at 37°C in a humidified atmosphere with 5% CO2.
-
a.
-
2.DAY 1 – Day of Transfection.
-
a.In a sterile tube, dilute total plasmid DNA (μg) in 500 μL diluent (HBSS). Use transfer vector (UbC-mNeonGreen-P2A-Akaluc): viral packaging (psPAX2): viral envelope (pDM2G) at 4:2:1 ratio (6:3:1.5 μg, respectively).
-
b.Add 42 μL of PEI (1 μg/μL) to the diluted DNA. Mix immediately by pipeting up and down / vortexing. The volume of PEI used is based on a 4:1 ratio of PEI (μg): total DNA (μg).
-
c.Incubate 10–15 min at 20°C–25°C.
-
d.Add 500 μL of DNA/PEI mixture to each plate of cells and incubate (37°C 24 h).
-
a.
-
3.DAY 2 – 18–24 h Post-Transfection.
-
a.Medium change to DMEM 5 mL + 10 μM Forskolin (5 mM stock: 10 μL) /dish.
-
b.Incubate (37°C 48 h).
-
a.
-
4.DAY 4 – Lentivirus collection.
-
a.Collect supernatant 48 h after transfection and filter through a 0.45 μm filter.
-
b.Concentrate virus by ultracentrifugation at 40,000 g, 2 h, 4°C.
-
c.After centrifugation, discard the supernatant carefully and resuspend the viral pellet in sterile PBS.
-
d.Aliquot the viral suspension and store at −80°C until use. Avoid repeated freeze–thaw cycles.
-
a.
Figure 1.
Lentiviral vector design and titration strategy
(A) Schematic map of the UBC-mNG-2A-Akaluc lentiviral vector. This construct is driven by the human UBC promoter and contains mNeonGreen (mNG) and Akaluc separated by a self-cleaving P2A peptide, allowing bicistronic expression.
(B) Representative flow cytometry plots of 293T cells transduced with 1 μL of lentivirus (left) or a negative control (right). mNG+ cells were detected using a FITC-compatible channel.
(C) Titration based on the percentage of mNG+ cells after transduction of 293T cells with increasing amounts of lentivirus. Data points in the linear range were used to calculate infectious units per milliliter (IU/mL). The line represents the fitted equation: Y = 53.82 × X.
Lentiviral titration
Timing: 4 days
-
5.
Seed 1 × 105 293T cells per well in a total of 11 wells of a 6-well plate using DMEM supplemented with 10% FBS and 1% PSG. Incubate the cells at 37°C in a humidified atmosphere with 5% CO2 overnight.
-
6.
On the following day, dilute the lentiviral stock in PBS to prepare 1:10 and 1:50 working solutions. Add the diluted virus to the pre-seeded 293T cells as follows:
From the 1:10 dilution: add 2, 5, 10, 20, or 50 μL per well.
From the 1:50 dilution: add 2, 5, 10, 20, or 50 μL per well.
Leave one well without virus as a negative control.
-
7.
After 48 h of incubation, harvest the cells from each well. Wash the cells with 10 mL of PBS and centrifuge at 1500 rpm for 5 min at 4°C.
-
8.
Resuspend the pellet in 500 μL of PBS containing PI at a final concentration of 1 μg/mL to exclude dead cells. Filter the suspension through a 40 μm cell strainer into a new tube.
-
9.
Analyze the percentage of mNG-positive cells in each sample by flow cytometry using a FITC channel. Any flow cytometer equipped with a FITC-compatible channel is suitable for this analysis (Figure 1B).
-
10.
Plot the percentage of mNG+ cells on the y-axis and the volume of virus added on the x-axis. Use the linear portion of the curve to calculate the viral titer in infectious units per milliliter (IU/mL) (Figure 1C).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse ckit-APC (2B8) (dilution 1/100) | Thermo Fisher Scientific | Cat#17-1171-83 ; RRID:AB_469431 |
| Anti-mouse CD4-Biotin (RM4-5) (dilution 1/2,000) | Thermo Fisher Scientific | Cat#13-0042-85; RRID:AB_466330 |
| Anti-mouse CD8a-Biotin (53–6.7) (dilution 1/2,000) | Thermo Fisher Scientific | Cat#13-0081-85; RRID:AB_466347 |
| Anti-mouse CD45R(B220)-Biotin (RA3-6B2) (dilution 1/500) | Thermo Fisher Scientific | Cat#13-0452-82; RRID:AB_466449 |
| Anti-mouse TER119-Biotin (TER119) (dilution 1/500) | Thermo Fisher Scientific | Cat#13-5921-85; RRID:AB_466798 |
| Anti-mouse Gr1-Biotin (RB6-8C5) (dilution 1/500) | Thermo Fisher Scientific | Cat#13-5931-85-85; RRID:AB_466801 |
| Anti-mouse CD11b-Biotin (M1/70) (dilution 1/100) | Thermo Fisher Scientific | Cat#13-0112-85; RRID:AB_466360 |
| Anti-mouse CD127-Biotin (A7R34) (dilution 1/500) | Thermo Fisher Scientific | Cat#13-1271-85; RRID:AB_466589 |
| Anti-mouse CD34-FITC (RAM34) (dilution 1/200) | Thermo Fisher Scientific | Cat#11-0341-85; RRID:AB_465022 |
| Anti-mouse Sca-1-PE (D7) (dilution 1/200) | Thermo Fisher Scientific | Cat#12-5981-83; RRID:AB_466087 |
| Anti-mouse CD201-PE (eBio1560) (dilution 1/200) | Thermo Fisher Scientific | Cat#12-2012-82; RRID:AB_914317 |
| Anti-mouse CD150-PE/Cy7 (TC15-12F12.2) (dilution 1/200) | BioLegend | Cat#115913; RRID:AB_439796 |
| Anti-mouse CD16/32-PE/Cy7(93) (dilution 1/200) | BioLegend | Cat#101318; RRID: AB_2104156 |
| Streptavidin-APC/eFluor780 (dilution 1/200) | Thermo Fisher Scientific | Cat#47-4317-82; RRID:AB_10366688 |
| Chemicals, peptides, and recombinant proteins | ||
| Polyvinyl alcohol (PVA), 87%–90% hydrolyzed | Sigma-Aldrich | Cat#P8136; CAS 9002-89-5 |
| Recombinant murine TPO | PeproTech | Cat#315-14; P40226 |
| Recombinant murine SCF | PeproTech | Cat#250-03; P20826 |
| Insulin-Transferrin-Selenium | Thermo Fisher Scientific | Cat#41400045 |
| Penicillin-Streptomycin-L-Glutamine solution (×100) | Wako | Cat#161-23201 |
| Propidium iodide | Sigma | Cat#P4170 |
| Forskolin | Wako | Cat#067-02191 |
| TokeOni (AkaLumine-HCI) | Kurogane Kasei/Wako | Cat#012-26701 |
| Recombinant DNA | ||
| Plasmid psPAX2 | Addgene | #12260 |
| Plasmid pMD2.G | Addgene | #12259 |
| Akaluc lentiviral vector | Yogo et al.1; Iwano et al.2 | Akaluc fragment was kindly provided by Dr. S. Iwano |
| Experimental models: Organisms/strains | ||
| Mouse: CD45.2+ C57BL/6: C57BL/6NCrSlc (8 weeks) | SLC | RRID:MGI:5295404 |
| Mouse: CD45.1+ C57BL/6: B6.SJL-Ptprca Pepcb/BoyJ (8 weeks) | Sankyo Labo | RRID:IMSR_JAX:002014 |
| Mouse: B6N-Tyrc-Brd/BrdCrCrl (B6 Albino) (8 weeks) | Charles River Laboratories | RRID:IMSR_CRL:493 |
| Other | ||
| Ham’s F-12 medium | Wako | Cat#087-08335 |
| D-MEM (high glucose) with L-glutamine, phenol red, and sodium pyruvate | Wako | Cat#043-30085 |
| HBSS(−) without phenol red | Wako | Cat#085-09355 |
| HEPES | Thermo Fisher Scientific | Cat#15630080 |
| Fetal bovine serum | Corning | Cat#35-079-CV |
| PBS(−) | Wako | Cat#166-23555 |
| Propidium iodide | Sigma-Aldrich | Cat#P4170 |
| MidiMACS Separator | Miltenyi Biotec | Cat#130-042-302 |
| LS columns | Miltenyi Biotec | Cat#130-042-401 |
| TPP tissue culture plates (96-well plate, round bottom) | Sigma-Aldrich | Cat#Z707899 |
| Falcon 15 mL High Clarity PP centrifuge tube | Corning | Cat#352196 |
| Falcon 50 mL High Clarity PP centrifuge tube | Corning | Cat#352070 |
Materials and equipment
Murine HSC expansion medium
| Reagent | Final concentration |
|---|---|
| Ham’s F-12 medium | – |
| HEPES | 10 mM |
| Murine SCF (PeproTech) | 10 ng/mL |
| Murine TPO (PeproTech) | 100 ng/mL |
| Polyvinyl alcohol (PVA, 84% hydrolyzed) | 0.1% |
| ITS supplement (Thermo Fisher) | 1× (1:100 dilution) |
| Penicillin-Streptomycin-Glutamine (PSG, Wako) | 1% |
Do not store as a complete mix. Store individual stock solutions at recommended conditions and assemble fresh medium for each experiment.
Lineage-Biotin antibody mix
| Antibody | PBS | |
|---|---|---|
| anti-mouse Gr1-Biotin | 100 μL | 0 μL |
| anti-mouse CD11b-Biotin | 50 μL | 50 μL |
| anti-mouse TER119-Biotin | 100 μL | 0 μL |
| anti-mouse CD4-Biotin | 25 μL | 75 μL |
| anti-mouse CD8a-Biotin | 25 μL | 75 μL |
| anti-mouse CD45R(B220)-Biotin | 50 μL | 50 μL |
| anti-mouse CD127-Biotin | 50 μL | 50 μL |
To prepare the Lineage antibody cocktail, the listed antibodies are mixed in the indicated volumes. The cocktail can be stored at 4°C for up to one week.
Step-by-step method details
Isolation and ex vivo culture of murine HSCs
Timing: 7 h
This step describes how to isolate murine hematopoietic stem cells (HSCs) from whole bone marrow (WBM) and how to culture them in serum-free medium prior to lentiviral transduction. CD34−CD150+KSL cells are used as the input population.
To facilitate clear distinction between donor-derived and host hematopoietic cells in downstream transplantation experiments, CD45.1 congenic mice are used as the source of donor HSCs, while CD45.2 mice serve as recipients. This congenic marker system enables reliable discrimination via flow cytometry.
-
1.Isolate whole bone marrow cells.
-
a.Sacrifice 8–10-week-old male C57BL/6-CD45.1 mice under isoflurane anesthesia in accordance with institutional animal care guidelines.
-
b.Excise the femurs, tibias, and pelvic bones.
-
c.Crush the bones in cold PBS using a sterile mortar and pestle.
-
d.Filter the cell suspension through a 40 μm strainer and count total nucleated cells, which are typically around 100 million per mouse.
-
a.
-
2.Enrich for c-Kit+ cells.
-
a.Centrifuge the cells at 440 × g for 5 min at 4°C and carefully aspirate the supernatant. Resuspend the cells in 200 μL of PBS.
-
b.Add 2 μL of anti-c-Kit-APC antibody and incubate for 30 min on ice.
-
c.Wash the cells by adding 10 mL of PBS, filter through a 40 μm strainer into a new 15 mL tube. Centrifuge for 5 min at 440 × g and 4°C.
-
d.Carefully aspirate the supernatant and resuspend the cells in 500 μL of PBS.
-
e.Add 2 μL of anti-APC magnetic microbeads and incubate for 15 min at 4°C.
-
f.Wash the cells again with 10 mL of PBS, filter into a new 15 mL tube. Centrifuge for 5 min at 440 × g and 4°C.
-
g.While centrifuging, prepare an LS column by placing it in a MidiMACS Separator and equilibrating with 2 mL of PBS. Place a 15 mL collection tube beneath the column.
-
h.After centrifugation, aspirate the supernatant and resuspend the cells in 2 mL of PBS. Apply the entire suspension to the LS column.
-
i.Wash the original tube with 3 mL of PBS and apply the wash to the LS column after the initial flow-through has passed. Repeat this washing step two more times, for a total of three washes.
-
j.Remove the LS column from the magnetic separator and place it on a new 15 mL tube. Elute the magnetically retained c-Kit+ cells by applying 5 mL of PBS and gently pushing the plunger through the column. Repeat with a second 5 mL of PBS to ensure complete elution.
-
k.Count the cells using an automated counter. Yields are typically 1–2 million c-Kit+ cells per mouse.
-
a.
-
3.Stain for HSC surface markers.
-
a.Centrifuge the c-Kit+ cells at 440 × g for 5 min at 4°C and carefully aspirate the supernatant.
-
b.Resuspend the pellet in 200 μL of PBS and add 3 μL of Lineage-Biotin antibody mix.
-
c.Incubate the cells for 30 min on ice.
-
d.Wash the cells with 10 mL of PBS, centrifuge at 440 × g for 5 min at 4°C and aspirate the supernatant.
-
e.Resuspend the pellet in 200 μL of PBS, and add the following antibodies (CD34-FITC, Sca-1-PE, c-Kit-APC, streptavidin-APC/eFluor, and CD150-PE/Cy7) at 1 μL for each antibody.
-
f.Incubate for 90 min on ice.
-
g.Wash the cells with 10 mL of PBS, filter through a 40 μm cell strainer into a new tube, and centrifuge at 440 × g for 5 min at 4°C.
-
h.Resuspend the cells in 300 μL of PBS containing PI (final concentration 1 μg/mL) to exclude dead cells.
-
a.
-
4.Prepare expansion plates before sorting.
-
a.Dispense 200 μL per well of HSC expansion medium3 that has been pre-warmed to 37°C into U-bottom 96-well plates.
-
b.Place the plates on ice.
-
a.
-
5.Sort HSCs by flow cytometry.
-
a.Use a BD FACSAria III cell sorter equipped with a 100 μm nozzle. Prior to running the sample, filter the cell suspension through a 40 μm mesh to avoid clogs.
-
b.Prepare compensation controls using mouse whole bone marrow cells stained individually with each fluorochrome-conjugated antibody, along with an unstained control. Perform compensation setup using these single-stained and non-stained samples.
-
c.Gate and sort the CD34−CD150+c-Kit+Sca-1+Lin− (CD34−CD150+KSL) population using the following gating strategy (Figure 2):
-
i.Exclude doublets by using FSC-A vs. FSC-H.
-
ii.Exclude dead cells by gating out PI-positive events.
-
iii.Plot Lin versus c-Kit and gate on Lin− cells.
-
iv.Plot c-Kit versus Sca-1 and gate on the c-Kit+Sca-1+ population.
-
v.Finally, plot CD150 versus CD34 and select the CD34−CD150+ fraction.For improved discrimination of CD34− cells, include an additional control sample that contains all antibodies except CD34-FITC; this will help define the negative boundary. Using the CD150 vs. CD34 plot further facilitates accurate gating of the CD34− population.
-
i.
-
d.Sort 100–200 cells per well directly into the pre-warmed U-bottom 96-well plates containing 200 μL of HSC expansion medium per well.
-
e.Immediately transfer the 96-well plates to a humidified CO2 incubator set to 37°C with 5% CO2.
-
f.Allow the sorted cells to recover and expand overnight (16–20 h) prior to proceeding to lentiviral transduction.
-
g.Allow cells to recover and expand overnight before proceeding to lentiviral transduction.
 CRITICAL: Although staining buffers containing fetal bovine serum (FBS) are commonly used in flow cytometry procedures, this protocol employs FBS-free PBS throughout all staining and washing steps. The use of serum-free conditions is essential to maintain the undifferentiated state of HSCs during and after sorting. Even minimal contamination with FBS can induce unwanted differentiation and negatively impact the subsequent ex vivo expansion of HSCs.
CRITICAL: Although staining buffers containing fetal bovine serum (FBS) are commonly used in flow cytometry procedures, this protocol employs FBS-free PBS throughout all staining and washing steps. The use of serum-free conditions is essential to maintain the undifferentiated state of HSCs during and after sorting. Even minimal contamination with FBS can induce unwanted differentiation and negatively impact the subsequent ex vivo expansion of HSCs.
-
a.
Figure 2.
Gating strategy for HSC purification
Representative flow cytometric plots showing the gating strategy for isolating HSCs from C57BL/6-CD45.1 (Ly5.1) mice. PI-negative cells are first gated on c-Kit and Lineage markers to select the Lineage− population. This population is then analyzed using Sca-1 and c-Kit expression, and the Sca-1+c-Kit+ subset is selected as shown. Finally, these cells are plotted based on CD34 and CD150 expression, and the CD34−CD150+ subset is gated. HSCs are defined as CD34−CD150+c-Kit+Sca-1+Lineage− (KSL) cells.
Lentiviral transduction and expansion of HSCs
Timing: 1 h
This step describes the transduction of HSC, which were freshly sorted one day prior, with a lentiviral vector encoding mNeonGreen-P2A-Akaluc under the UbC promoter, followed by ex vivo expansion under serum-free HSC expansion medium.
-
6.Add lentivirus to cultured HSCs.
-
a.Thaw a frozen aliquot of concentrated lentiviral vector on ice.
-
b.Add virus to each well of cultured HSCs at a multiplicity of infection (MOI) of 300.
-
a.
-
7.Incubate transduced cells.
-
a.Return the plate to the CO2 incubator (37°C, 5% CO2).
-
b.Incubate for 24 h.
-
a.
-
8.Replace half of the medium.
-
a.After 24 h, gently remove 100 μL of medium from each well.
-
b.Add 100 μL of fresh pre-warmed HSC expansion medium to each well.
-
c.Repeat this half-medium change again on the following day.
-
a.
-
9.Continue ex vivo expansion.
-
a.Maintain cultures in a humidified incubator at 37°C, 5% CO2 for a total of 7 days.
-
b.Perform medium changes every 2–3 days.
-
c.Monitor transduction efficiency by mNeonGreen (mNG) fluorescence using flow cytometry if necessary (excitation: 506 nm; emission: 517 nm).
-
a.
CRITICAL: Avoid FBS contamination from virus preparation. Residual serum can promote unwanted differentiation of HSCs. Medium exchange is critical after viral transduction.
FACS sorting of expanded HSC and progenitor populations
Timing: 3 h
This step describes how to isolate specific hematopoietic stem and progenitor cell populations from expanded cultures using flow cytometry.
-
10.Harvest expanded cells for sorting.
-
a.Collect cultured cells by pipetting the contents of each well.
-
b.Pool cells if needed and transfer to 1.5 mL tubes.
-
c.Wash cells with PBS and centrifuge at 440 × g for 5 min at 4°C.
-
d.Discard the supernatant and resuspend the pellet in fresh PBS.
-
a.
-
11.Stain cells for surface markers.
-
a.Resuspend the cultured bulk cells in 200 μL of PBS and add 1 μL of Lineage-Biotin antibody mix. Incubate the cells on ice for 30 min.
-
b.Wash the cells by adding 10 mL of PBS and centrifuge at 1500 rpm for 5 min at 4°C. Aspirate the supernatant.
-
c.Resuspend the cell pellet in 200 μL of PBS and add the following antibodies at 1 μL: Streptavidin-BV421, c-Kit-APC, Sca-1-APC/Cy7, CD150-PE/Cy7, and CD201-PE. Incubate on ice for 30 min.
-
d.Wash the cells again with 10 mL of PBS and centrifuge at 440 × g for 5 min at 4°C. Aspirate the supernatant and resuspend the cells in an appropriate volume of PBS.
-
e.Just before sorting, add PI to the cell suspension at a final concentration of 1 μg/mL to exclude dead cells.
-
a.
-
12.Sort expanded HSCs by flow cytometry.
-
a.Use a BD FACSAria III with a 100 μm nozzle and appropriate filters.Gate and sort mNeonGreen+CD201+CD150+c-Kit+Sca-1+Lineage− (Akaluc+ expanded HSCs) using the following gating strategy (Figure 3).
-
i.Exclude doublets by using FSC-A vs. FSC-H.
-
ii.Exclude dead cells by gating out PI-positive events.
-
iii.From the PI− population, select mNG+ cells.
-
iv.Plot mNG+ cells for c-Kit versus Lineage markers, and gate on the Lineage− population.
-
v.Plot the Lineage− cells for Sca-1 versus c-Kit, and gate on the Sca-1+c-Kit+ population.
-
vi.Finally, plot the Sca-1+c-Kit+ cells for CD201 versus CD150 expression, and gate on the CD201+CD150+ subset.Ex vivo expanded murine HSCs uniformly upregulate CD34 expression, which alters their surface marker profile compared to freshly isolated HSCs. As previously reported,4,5 functionally potent HSCs after ex vivo culture are enriched in the CD201+CD150+c-Kit+Sca-1+Lineage− population. Therefore, a distinct gating strategy is used for expanded HSCs, which differs from the conventional CD34−CD150+KSL strategy employed for freshly isolated HSCs.
-
i.
-
b.Collect sorted cells into tubes containing HSC expansion medium and keep on ice until transplantation.Note: Perform all staining steps at 4°C or on ice. Use serum-free PBS throughout to preserve stem cell properties.
-
a.
Figure 3.
Gating strategy for Akaluc+ expanded HSCs
Representative flow cytometric plots of ex vivo expanded HSCs after 7 days of culture.
PI-negative cells are first gated for mNeonGreen (mNG) positivity. The mNG+ cells are then plotted for c-Kit and Lineage markers, and the Lineage− population is selected. This population is further analyzed based on Sca-1 and c-Kit expression, and Sca-1+c-Kit+ cells are gated. Finally, these cells are plotted for CD201 and CD150 expression, and the CD201+CD150+ subset is selected. Akaluc+ expanded HSCs are defined as mNeonGreen+CD201+CD150+c-Kit+Sca-1+Lineage− cells.
Transplantation into irradiated recipient mice
Timing: 1–2 h (day 7)
This step describes the intravenous transplantation of expanded and sorted HSCs into lethally irradiated recipient mice. To support hematopoietic recovery and ensure the survival of recipients, we co-transfer 5 × 105 whole bone marrow (WBM) cells derived from C57BL/6-CD45.1/CD45.2 F1 mice as competitor or support cells. These F1 cells co-express both CD45.1 and CD45.2, enabling clear distinction from CD45.1+ donor HSCs and CD45.2+ recipient cells by flow cytometry. This setup also allows for accurate assessment of donor-derived reconstitution.
-
13.Prepare recipient mice.
-
a.Use 8–12-week-old C57BL/6-CD45.2 mice as recipients.
-
b.Irradiate mice with 8.0 Gy total body irradiation. X-ray irradiation is performed using the MBR-1618R-BE system (Hitachi Power Solutions).
-
a.
-
14.Prepare donor cell suspension.
-
a.Mix sorted expanded HSCs with 5 × 105 whole bone marrow cells from C57BL/6-CD45.1/CD45.2 F1 mice as competitor/support cells.
-
b.Keep the cell suspension on ice until injection.
-
a.
-
15.Perform transplantation.
-
a.Inject the cell suspension intravenously via the tail vein using a 27G needle.
-
b.Return mice to cages and monitor daily for recovery.
-
a.
Note: Transplantation must be completed on the same day as sorting. Work quickly and maintain sterility throughout.
In vivo bioluminescence imaging
Timing: 2 h (daily imaging)
This step describes noninvasive tracking of hematopoietic reconstitution dynamics in recipient mice using the AkaBLI system. Imaging is performed daily from Day 5 to Day 20 using intraperitoneal injection of the Akaluc substrate, TokeOni.
-
16.Prepare for imaging.
-
a.Use an IVIS Lumina Series III or IVIS Spectrum imaging system (PerkinElmer).
-
b.Ensure that the system is pre-warmed and that the gas lines for isoflurane anesthesia are functioning properly.
-
c.Prepare fresh aliquots of 15 mM TokeOni substrate prior to imaging.
-
a.
-
17.Inject luciferin substrate.
-
a.Inject 50 μL of undiluted 15 mM TokeOni intraperitoneally into each mouse.
-
b.Wait 5 min post-injection before imaging.
-
c.Anesthetize mice with isoflurane and place them on the imaging stage.
-
a.
-
18.Capture images.
-
a.Acquire 2D luminescence images.
-
b.Set the exposure time to 60 s and binning to medium. If the signal is weak, extend the exposure time as needed. If the signal is strong and a saturation warning appears, shorten the exposure time accordingly.
-
c.Set the field of view (FOV) to “D,” which allows simultaneous imaging of five mice: place a control mouse (injected with TokeOni only) on the far left, and four experimental mice to its right.
-
d.Repeat imaging daily from Day 5 to Day 20 post-transplantation.
-
a.
-
19.Analyze signal intensity.
-
a.Open images using Living Image Software.
-
b.Define a fixed region of interest (ROI) over the spleen or bone marrow region for each mouse using the ROI tools.
-
c.Select “Avg Radiance (p/s/cm2/sr)” to calculate the average radiance for each ROI across time points.
-
d.Auto-luminescence may occur, particularly around the liver region. To account for this, draw the same type of ROI on negative control animals (e.g., mice injected with TokeOni only), and obtain their average radiance. Subtract this background signal from the average radiance of experimental samples to improve accuracy.
-
e.Compare signal kinetics across different experimental groups.
-
a.
Note: Autoluminescence may occur, especially in the liver. Include negative controls (non-transduced HSC-transplanted mice) in every imaging session. Perform imaging under consistent conditions for longitudinal comparison.
Expected outcomes
Representative flow cytometry plots for the isolation of HSCs from mouse bone marrow are shown in Figure 2. Following isolation, HSCs are transduced with a lentiviral vector encoding the Akaluc reporter gene. A typical flow cytometric profile of Akaluc-transduced HSCs is shown in Figure 3, where mNG+ cells represent successfully labeled populations. Under optimized conditions as described in this protocol, transduction efficiency typically reaches 50%–70%, allowing for robust downstream imaging.
Transplantation of 3,000 mNG+CD201+CD150+c-Kit+Sca-1+Lin− (Akaluc+ Expanded HSCs) into lethally irradiated B6 Albino recipient mice enables the in vivo longitudinal visualization of HSC-derived hematopoietic activity via bioluminescence imaging (Figure 5). Robust signals are consistently observed in the spleen and bone marrow regions. To confirm successful multilineage engraftment, we performed flow cytometric analysis of peripheral blood at later time points. As shown in Figure 4, Akaluc+ HSCs gave rise to Gr-1+Mac-1+ myeloid cells, B220+ B cells, and CD4+CD8+ T cells, demonstrating that the bioluminescent signal is inherited by downstream progeny. This result validates that the transplanted Akaluc+ HSCs maintain their multipotency and contribute to major hematopoietic lineages in vivo.
Figure 5.
Spatiotemporal dynamics of transplanted HSCs visualized by AkaBLI
(A) Representative IVIS images of recipient B6 Albino mice transplanted with 3,000 Akaluc+CD201+CD150+KSL cells following 8 Gy total body irradiation. Mice were injected intraperitoneally with 50 μL of 15 mM TokeOni daily, and bioluminescence imaging was performed 5 min post-injection, starting from Day 5 post-transplant. The color scale indicates radiance intensity ranging from 1 × 106 to 3 × 107 photons/sec/cm2/sr. Lower intensities appear in blue/purple, while higher intensities appear in yellow/red.
(B–D) Quantitative analysis of radiance over time. Regions of interest (ROIs) were placed over the whole body (B), spleen (C), and skull (D), and the mean luminescence intensity was plotted over time to asses HSC-derived hematopoietic activity.
Figure 4.
Peripheral blood chimerism after transplantation of Akaluc-labeled HSCs
Akaluc+CD150+CD201+KSL cells (n = 5) were transplanted at 3,000 cells per mouse, resulting in long-term engraftment. Donor-derived Gr-1+/Mac-1+ myeloid cells, B220+ B cells, and CD4+/CD8+ T cells were detected in the peripheral blood, confirming multilineage differentiation. Error bars represent SD.
Using ROI-based analysis in Living Image Software, changes in mean luminescence intensity over time can be quantitatively evaluated (Figure 5). Notably, the bioluminescence signal in the spleen exhibits a characteristic multiphasic pattern, including transient decreases around days 10 and 13 post-transplantation. This is not due to technical variability but rather reflects a biologically relevant phenomenon. In our previous study,1 we demonstrated that post-transplant hematopoiesis proceeds through a spleen-dominant “hematopoietic cell inflation” process characterized by three distinct hematopoietic peaks. Thus, the observed signal fluctuation likely corresponds to dynamic biological events in the spleen during early hematopoietic reconstitution.
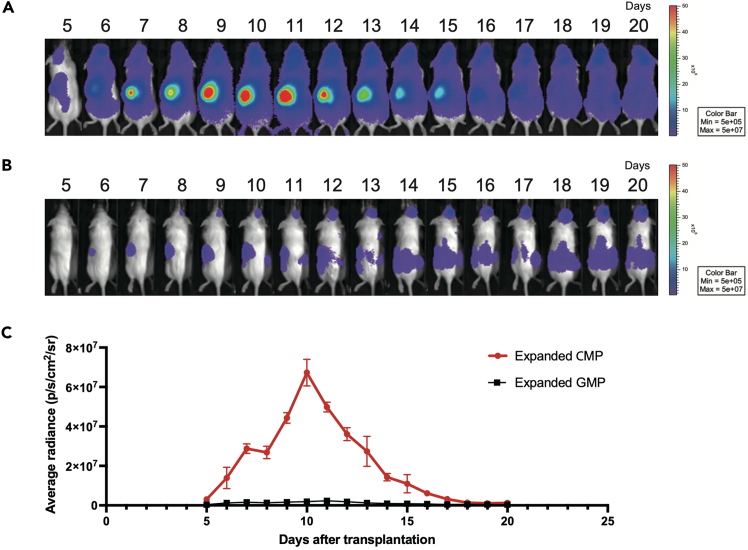
Furthermore, by varying the donor cell populations, this protocol enables comparative analysis of hematopoietic dynamics among different progenitor subsets.1 For example, transplantation of 10,000 mNG+c-Kit+Sca-1−CD34+FcγR−Lin− (expanded CMPs) versus 10,000 mNG+c-Kit+Sca-1−CD34+FcγR+Lin− (expanded GMPs) results in markedly different temporal patterns of signal intensity and tissue distribution (Figure 6), indicating their distinct in vivo behaviors.
Figure 6.
Post-transplant dynamics of distinct progenitor populations
(A and B) Representative bioluminescence images of mice transplanted with 10,000 Akaluc+ expanded CMPs (A) or GMPs (B) after 7 days of culture. Cells were transplanted into lethally irradiated B6 Albino mice (8 Gy), and luminescence signals were acquired daily following intraperitoneal TokeOni injection. The color scale indicates radiance intensity ranging from 5 × 105 to 5 × 107 photons/sec/cm2/sr. Lower intensities appear in blue/purple, while higher intensities appear in yellow/red.
(C) Quantification of mean radiance in the spleen ROI over time. Temporal dynamics of spleen-localized signals were compared between expanded CMPs and GMPs. Data are presented as mean values with standard deviation (SD); n=3 mice per group.
Altogether, this protocol enables sensitive, noninvasive, and quantitative assessment of hematopoietic stem and progenitor cell derived hematopoietic reconstitution in live mice over time.
Limitations
This protocol utilizes the Akaluc bioluminescence reporter system driven by the UbC promoter, which enables robust and stable expression across all hematopoietic lineages. However, because the promoter is non-lineage-specific, this system does not permit the discrimination of downstream progenitor cells after HSC differentiation. Once HSCs give rise to mature blood cells, all daughter lineages express the reporter equally, contributing collectively to the detected bioluminescence signal. Consequently, this method is not suitable for lineage-restricted tracking. To overcome this limitation, future applications may incorporate lineage-specific promoters to restrict reporter expression or utilize spectrally distinct luciferases for multiplexed imaging of different hematopoietic subsets.
Troubleshooting
Problem 1
Weak or undetectable luminescence signals.
Potential solution
This may result from insufficient Akaluc expression or transplantation of too few cells. Ensure high-quality lentiviral preparation and confirm viral titer using a transduction-based assay (see lentiviral titration). Avoid serum contamination during virus production. If only a small number of HSCs is transplanted, the bioluminescence signal may take 2–3 days to reach detectable levels due to in vivo expansion. For early imaging (e.g., Day 1 post-transplant), transplant a larger number of cells (e.g., 10,000 cells) to ensure detectable signal intensity.
Problem 2
High background signal in the liver.
Potential solution
TokeOni (Akalumine) causes low-level auto-luminescence, especially in the liver. This can mask weak, biologically relevant signals from liver-homing HSCs. Always include negative control mice to distinguish background from true signal. To detect liver-localized hematopoietic activity, it may be necessary to transplant a larger number of labeled cells to exceed the background threshold (see in vivo bioluminescence imaging).
Problem 3
Low transduction efficiency of HSCs
Potential solution
Lentivirus transduction may be inefficient in primitive HSCs if the virus titer is too low, or the vector quality is poor (see Preparation of Akaluc lentiviral vector). Concentrate lentivirus using ultracentrifugation. Avoid serum contamination, which can promote differentiation. Ensure that CD34−CD150+KSL cells are used as the transduction target and verify transgene expression post-transduction by flow cytometry.
Problem 4
Poor yield of target population during sorting.
Potential solution
Undesired differentiation during culture may reduce the yield of phenotypically defined HSCs. Even minimal serum contamination from FBS in virus stocks can lead to HSC differentiation. Perform half-medium changes after transduction to remove residual virus and any serum components. For more stringent HSC gating, consider including makers such as CD48 to further enrich for stem cell populations.
Alternatives: While PVA-based serum-free culture supports efficient HSC expansion, our laboratory has demonstrated that using an alternative polymer, Soluplus (BASF), enables comparable or even enhanced expansion of functional HSCs.4
Problem 5
Variable signal kinetics among mice.
Potential solution
Functional heterogeneity in the transplanted HSC population may lead to inconsistent bioluminescence kinetics. Although CD201+CD150+KSL cells are highly enriched for HSCs, minor progenitor contamination can cause variability. To reduce this, increase the transplanted cell number or incorporate additional markers to refine the sorted population.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Satoshi Yamazaki (y-sato4@g.ecc.u-tokyo.ac.jp).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Takao Yogo (takayogo0430@g.ecc.u-tokyo.ac.jp).
Materials availability
Materials used in this study are available from the lead contact upon reasonable request.
Data and code availability
The published article includes all datasets generated or analyzed during this study. For additional access, the lead contact can be approached to request the data and materials. This protocol does not report original code.
Acknowledgments
We thank the University of Tokyo Institute of Medical Science (IMSUT) FACS core laboratory for technical assistance. Pictograms and illustrations were generated with BioRender (https://www.biorender.com/). This work was supported by the Japan Society for the Promotion of Science (JSPS; 24K02478, 21F21108, and 20K21612 to S.Y. and 24K19192 to T.Y.) and the Japan Agency for Medical Research and Development (AMED; 24bm1223011h0002, 23bm1223011h0001, 21bm0404077h0001, and 21bm0704055h0002 to S.Y. and JP25bm1123084 to T.Y.).
Author contributions
Conceptualization, T.Y. and S.Y.; methodology, T.Y. and S.Y.; investigation, T.Y.; analysis, T.Y.; writing, T.Y.; funding acquisition, T.Y. and S.Y.; supervision, S.Y.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Takao Yogo, Email: takayogo0430@g.ecc.u-tokyo.ac.jp.
Satoshi Yamazaki, Email: y-sato4@g.ecc.u-tokyo.ac.jp.
References
- 1.Yogo T., Becker H.J., Kimura T., Iwano S., Kuchimaru T., Miyawaki A., Yokomizo T., Suda T., Iwama A., Yamazaki S. Progenitor effect in the spleen drives early recovery via universal hematopoietic cell inflation. Cell Rep. 2025;44 doi: 10.1016/j.celrep.2025.115241. [DOI] [PubMed] [Google Scholar]
- 2.Iwano S., Sugiyama M., Hama H., Watakabe A., Hasegawa N., Kuchimaru T., Tanaka K.Z., Takahashi M., Ishida Y., Hata J., et al. Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science. 2018;359:935–939. doi: 10.1126/science.aaq1067. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson A.C., Ishida R., Kikuchi M., Sudo K., Morita M., Crisostomo R.V., Yamamoto R., Loh K.M., Nakamura Y., Watanabe M., et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. 2019;571:117–121. doi: 10.1038/s41586-019-1244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker H.J., Ishida R., Wilkinson A.C., Kimura T., Lee M.S.J., Coban C., Ota Y., Tanaka Y., Roskamp M., Sano T., et al. Controlling genetic heterogeneity in gene-edited hematopoietic stem cells by single-cell expansion. Cell Stem Cell. 2023;30:987–1000.e8. doi: 10.1016/j.stem.2023.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yogo T., Iwamoto Y., Becker H.J., Kimura T., Ishida R., Sugiyama-Finnis A., Yokomizo T., Suda T., Ota S., Yamazaki S. Quantitative phase imaging with temporal kinetics predicts hematopoietic stem cell diversity. Nat. Commun. 2025;16:6496. doi: 10.1038/s41467-025-61846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. For additional access, the lead contact can be approached to request the data and materials. This protocol does not report original code.