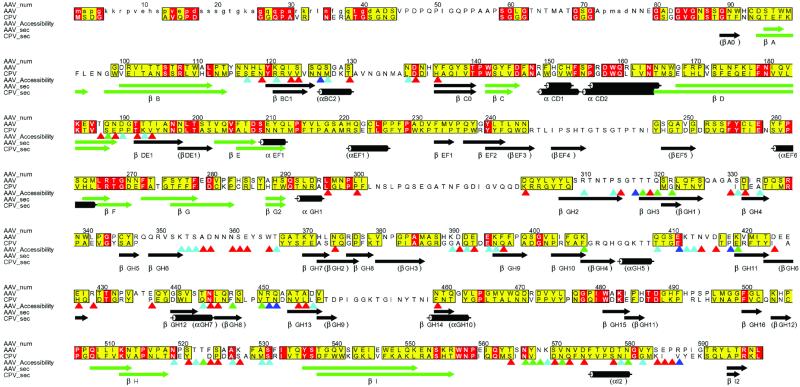
Fig 2.
Conservation of sequence and secondary structure. Sequences of AAV-2 and CPV were aligned according to structure, then improved in loop regions according to sequence [Programs STAMP, PILEUP, and ALSCRIPT (62–66)]. Residues that are identical in AAV-2 and CPV are red. Those with an ALSCRIPT similarity score >5 (measured from 0 to 10) are yellow. Secondary structures of AAV-2 and CPV are compared, the core β-barrel highlighted in green, and labeled according to AAV-2 (CPV in parentheses where different). Side chain (outer) surface accessibility was calculated with a 1.5 Å radius probe, and is shown with triangles shaded according to each residue's accessibility: 20 Å2 < red <50 Å2 < cyan <80 Å2 < green <100 Å2 < blue.