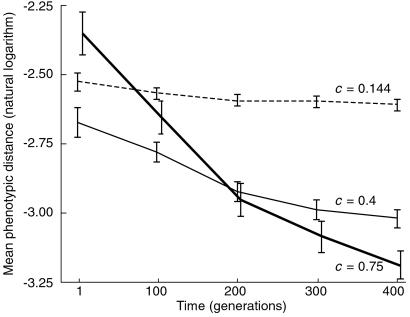
Fig 4.
Effect of network complexity on sensitivity to mutation. Three sets of 100 simulations were performed, with different average proportions of nonzero regulatory interactions: c = 0.75 (thick solid line), 0.4 (thin solid line), and 0.144 (broken line). Sensitivity to mutation was determined every 100 generations. Log-transformed values were analyzed, as untransformed data departed significantly from normality, whereas transformed data did not (Lilliefors tests). Plotted are mean sensitivities, expressed as mean phenotypic distances on perturbation; error bars are ±1 SEM. Two-way ANOVA on the transformed data yields p ≈ 0 for each main effect (c and time) as well as their interaction. Nonparametric Kruskal–Wallis tests on untransformed data confirm these strong effects.