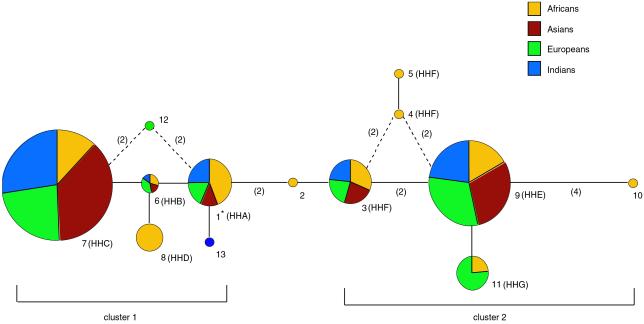
Fig 3.
A reduced median network of the 13 unique CCR5 5′ cis-regulatory region haplotypes found in the Old World panel. Haplotypes are also labeled according to the nomenclature we reported previously (refs. 11 and 12, and see Table 2). The size of each node is proportional to the haplotype frequency in the Old World panel, and the frequency of each haplotype within each continental population is indicated by the varied colors within each node. Branch lengths represent one nucleotide substitution, except where noted in parentheses. Dashed lines indicate alternate topologies of equal length. Haplotype 1, indicated with an asterisk, had the highest number of ancestral character states, differing from the chimpanzee consensus sequence by 7 nucleotide sites. This finding suggests that it may be the oldest lineage in the human sample and the root of the genealogy. It is also more common in Africans than any other continental population (Table 2), it is the most frequent haplotype in Africans, and it reaches a peak frequency of more than 70% (n = 68) in Mbuti Pygmies (11). Fifty percent of the CCR5 haplotypes in Asians are within 2 mutation steps of haplotype 1. Thus, although the CCR5 haplotypes in Africans are the most diverse and closest to the root, there is a substantial amount of haplotype diversity in Asians that is close to the root as well.