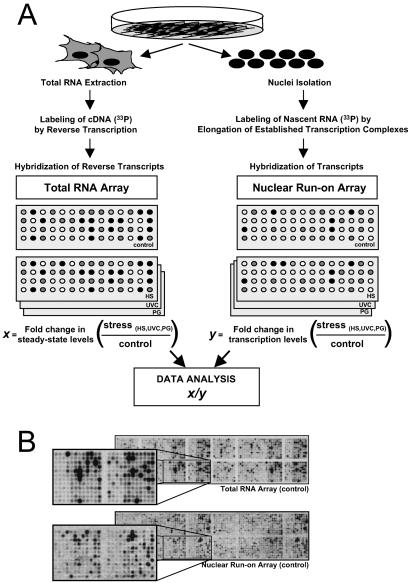
Fig 1.
Strategy to study stress-triggered alterations in mRNA turnover by using cDNA arrays. (A) Following treatment of H1299 cells with either UVC (20 J/m2), HS (43°C, 2 h), or PG (36 μM), two sets of radiolabeled probes were prepared: total RNA (Left), isolated 8 h after stimulation, was used to prepare radiolabeled cDNA through reverse transcription in the presence of [α-33P]dCTP; and newly transcribed RNA (Right), prepared 3 h after stimulation, was radiolabeled in a nuclear run-on reaction in the presence of [α-33P]UTP. x, fold difference in signal intensity (stress-treated relative to untreated) for a given gene on total RNA arrays; y, fold difference in signal intensity (stress-treated relative to untreated) for a given gene on nuclear run-on arrays. (B) Representative cDNA arrays to illustrate the hybridization signals corresponding to either steady-state RNA levels (total RNA array) or newly transcribed RNA (nuclear run-on array), in untreated populations. Correlation coefficients serving to evaluate the internal reproducibility of the signals on total RNA arrays and nuclear run-on arrays were 0.99 in each case (not shown). The array (ref. 8; www.grc.nia.nih.gov/branches/rrb/dna/array.htm) includes cDNA segments typically comprising the 3′ UTR and coding region. cDNA fragments range from 500 to 2,000 bp.