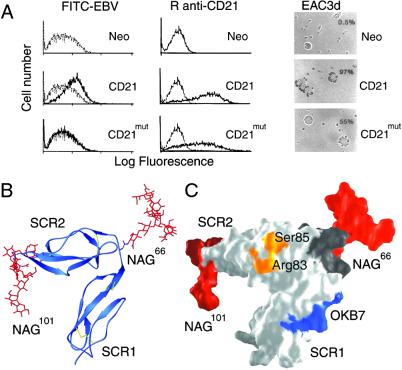
Fig 5.
Functional and structural characterization of a hCD21 glycosylation mutant, CD21mut. Mutation to Asn-66 causes glycosylation at this position as occurs naturally in mCD21, which does not bind EBV. (A) Comparison of ligand binding. Shown is binding of FITC-labeled EBV (FITC-EBV, Left), monospecific rabbit anti-CD21 antibody (R anti-CD21, Center), and C3d (EAC3d, Right) to L cells transfected with control vector (Neo), CD21, or CD21mut. (Left) Flow cytometry showing binding of FITC-EBV (dark line) to L cells transfected with CD21 but not to cells transfected with CD21mut or the control vector Neo. FITC-labeled avidin (gray line) was used as a control. (Center) Flow cytometry showing binding of R anti-CD21 antibody to CD21 and CD21mut (dark lines) but not to L cells transfected with vector alone. Preimmunization rabbit serum (gray line) was used as a control. (Right) Photomicrographs (magnification ×137) showing EAC3d binding to CD21 and CD21mut, but not to L cells transfected with vector alone. The percentage of cells binding >4 EAC3d in a field of 200 cells is indicated in the upper right corner and coordinates with receptor distribution and density on the respective transfectants. (B) Ribbon drawing of hCD21 SCR1–SCR2 with modeled murine glycosylation pattern. mCD21 has glycans at positions Asn-66 and Asn-101, and a large structurally known glycan structure (red; glycan-2 from Fig. 3A) was modeled onto each of these amino acids (see Methods). (C) Surface representation of the structure shown in B. The primary C3d-binding residues Arg-83 and Ser-85 (see Fig. 4A) are shown in orange, the OKB7 epitope (residues 8–15) is in blue, the linker region (residues 63–70) is in dark gray, and the two glycans are shown in red. The glycans are distant from the C3d binding site and most likely prevent EBV and OKB7 binding by steric hindrance.