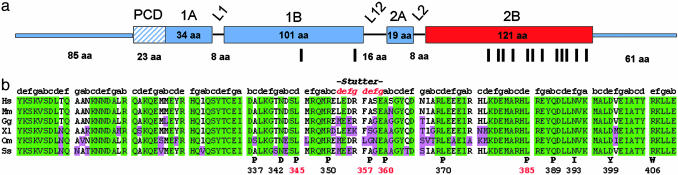
Fig. 1.
Desmin mutations found in the α-helical rod of human desmin. (a) Schematic view of the organization of the human desmin molecule. The α-helical central rod domain is interrupted by the three non-α-helical linker regions L1, L12, and L2, which results in the formation of four α-helical segments, termed coils 1A, 1B, 2A, and 2B. The N-terminal “head” and C-terminal “tail” segments are non-α-helical. PCD, precoiled-coil domain; red, region of the molecule where most mutations are located, i.e., coil 2B. Vertical bars depict the localization of all mutations investigated. (b) Comparison of the amino acid sequence of coil 2B of various desmin proteins (Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; XL, Xenopus laevis; Om, Oncorhynchus mykiss; Ss, Scyliorhinus stellaris; the alignment was performed by using clustal). Missense mutations of desmin are depicted in the bottom line. Green, all amino acids of a column are identical; purple, amino acid in one column differs from the corresponding amino acid in Hs. Note that the sequence of murine desmin is virtually identical to the human sequence (>99%), and that most mutations reside in highly conserved regions of the last two-thirds of coil 2B. Data for mutations highlighted red are presented in Fig. 2.