Abstract
White lupin (Lupinus albus) grown under P deficiency displays a suite of highly coordinated adaptive responses. Included among these is secretion of copious amounts of acid phosphatase (APase). Although numerous reports document that plants secrete APases in response to P deficiency, little is known of the biochemical and molecular events involved in this process. Here we characterize the secreted APase protein, cDNA, and gene from white lupin. The secreted APase enzyme is a glycoprotein with broad substrate specificity. It is synthesized as a preprotein with a deduced Mr of 52,000 containing a 31-amino acid presequence. Analysis of the presequence predicts that the protein is targeted to outside the cell. The processed protein has a predicted Mr of 49,000 but migrates as a protein with Mr of 70,000 on sodium dodecyl sulfate gels. This is likely due to glycosylation. Enhanced expression is fairly specific to proteoid roots of P-stressed plants and involves enhanced synthesis of both enzyme protein and mRNA. Secreted APase appears to be encoded by a single gene containing seven exons interrupted by six introns. The 5′-upstream putative promoter of the white lupin-secreted APase contains a 50-base pair region having 72% identity to an Arabidopsis APase promoter that is responsive to P deficiency. The white lupin-secreted APase promoter and targeting sequence may be useful tools for genetically engineering important proteins from plant roots.
P is a conundrum in agriculture and agroecosystems. While it is a critical macronutrient required for a myriad of functions in plants, P is also among the most limiting factors for plant growth due to its rapid immobilization by soil organic and inorganic components (Runge-Metzger, 1995; Von Uexkull and Mutert, 1995). Thus, large amounts of P fertilizer are applied to cropland in the developed world. This practice is not only expensive but is also polluting and nonsustainable. Abelson (1999) has noted that lack of inexpensive P is a potential future crisis in agriculture. Thus, it is imperative that we develop a fundamental understanding of the adaptive strategies among plants for acquiring adequate P in a nutrient-limited environment.
Anywhere from 30% to 80% of soil P occurs in organic complexes (Bieleski, 1973). While the overall importance of soil organic P in plant nutrition is unresolved, plants can utilize a number of P sources in both sand and soil culture (Duff et al., 1994). Utilization of soil organic P requires hydrolysis by phosphatases. P acquisition in many plants is thought to involve enhanced expression and secretion of acid phosphatase (orthophosphoric monoester phosphohydrolyases; EC 3.1.3.2). Under P-deficient conditions, acid phosphatases (APases) hydrolyze monoester soil organic P at low pH, thereby increasing orthophosphate availability (Tarafdar and Claasen, 1988; Duff et al., 1994). Enhanced synthesis and excretion of APase under P-deficient conditions has been documented in a number of plants (Tarafdar and Claasen, 1988; Goldstein, 1992; Wasaki et al., 1999, Haran et al., 2000). However, the biochemical and molecular elements affecting secretion of APase into the rhizosphere have received scant attention. Haran et al. (2000) reported the isolation of a 1,500-bp PCR fragment containing a portion of the promoter for an Arabidopsis membrane APase that was responsive to P deficiency. A chimeric gene containing 1,300 bp of APase promoter and the APase signal peptide fused to green fluorescent protein was used to transform Arabidopsis. Transformed plants when subjected to P deficiency secreted small amounts of green fluorescent protein into the rhizosphere.
White lupin (Lupinus albus) is a nitrogen-fixing legume highly efficient at acquiring soil P that is unavailable for other plants despite its lack of a mycorrhizal symbiosis (Neumann et al., 1999; Watt and Evans, 1999). Instead it has a suite of adaptations to P deficiency that facilitate efficient P acquisition, including formation of proteoid roots (cluster roots) (Johnson et al., 1996; Neumann et al., 1999), modified carbon metabolism to bypass P-requiring steps (Johnson et al., 1996), secretion of substantial amounts of citrate and malate from proteoid root zones, and secretion of copious amounts of a novel P deficiency-induced APase into the rhizosphere of proteoid roots (Ozawa et al., 1995; Gilbert et al., 1999).
An APase secreted from roots of P-deficient white lupin was purified by Tadano's group (Ozawa et al., 1995; Li and Tadano, 1996). In a subsequent follow-up study, Wasaki et al. (1999) isolated and sequenced a very unusual APase cDNA isolated from a cDNA library prepared with RNA from P-deficient white lupin roots. This lupin APase (LASAP1) cDNA is some 380 bp and 175 amino acids longer than other previously reported plant APases (Schenk et al., 2000) and contains a putative signal sequence of 31 amino acids that predict a plasma membrane location. However, the relationship of LASAP1 to exudation of APase activity from proteoid roots was not addressed. Neither was LASAP1 transcript accumulation related to development of proteoid roots. Our laboratory has documented (Gilbert et al., 1999) the secretion of large quantities of a novel APase from proteoid roots of P-deficient white lupin. This novel isoform of APase was not detectable in either roots or root exudates of P-sufficient plants and appears to be different than LASAP1 (S.S. Miller and C.P. Vance, unpublished data).
The objectives of this research were to isolate and produce antibodies to the APase secreted into the rhizosphere of proteoid roots of P-deficient white lupin, characterize the cDNA encoding the secreted APase, and assess transcript accumulation in proteoid and normal roots of P-sufficient versus P-deficient plants. Lastly, we were interested in isolating the white lupin-secreted APase gene and defining the sequence of the promoter.
RESULTS
Purification and Characterization of the Secreted APase Protein from Proteoid Root Exudate
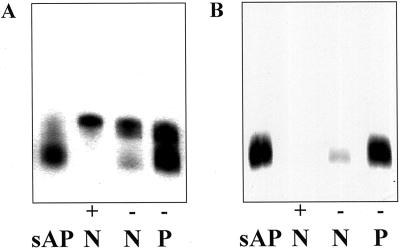
Previous results from our laboratory on APase isozyme expression revealed that a novel APase enzyme form (called isoform 2) was highly expressed by 14 days after emergence (DAE) in both −P proteoid and normal root cell-free extracts (Gilbert et al., 1999). Another isozyme (called isoform 1) appeared to be more ubiquitous in expression, being found in cell-free extracts from both +P and −P normal and proteoid roots. Isoform 2 was found to be the major APase form in −P proteoid roots and root exudates. Copious amounts of this novel APase were secreted into the rhizosphere of 14-DAE −P proteoid roots (Gilbert et al., 1999). We made use of the intrinsic secretion system to collect and purify the secreted APase (sAPase) and, since few proteins are secreted, purification was quickly accomplished via (NH4)2SO4 fractionation, gel electrophoresis, and subsequent electroelution. The enzyme was followed through the purification by in vitro enzyme assay and finally visualized at the last purification step by activity staining on polyacrylamide gels. Typical yields of total protein and APase enzyme activity from the initial exudation step were 35 μg of protein and 52 μmol of P liberated per hour, respectively, per gram fresh weight of proteoid root tissue. Sufficient quantities of enzyme were purified to allow for production of high titer antiserum in rabbits. The antiserum raised was able to immunoprecipitate at least 60% of the APase enzyme activity from an aliquot of partially purified sAPase (data not shown). In order to confirm the identity of the exuded, purified protein as isoform 2, the purified enzyme was electrophoresed on native-PAGE gradient gels alongside cell-free extracts made from normal (+P or −P) and proteoid (−P) roots, and the gels were stained for APase activity. The isoform pattern for P-sufficient and P-deficient root extracts was confirmed as previously published (Fig. 1A; Gilbert et al., 1999). As predicted, the purified enzyme migrated with isoform 2 (Fig. 1A), demonstrating that isoform 2 represents the secreted form of the enzyme found in P-deficient white lupin roots.
Figure 1.
Antibodies raised to white lupin sAPase are specific for the native enzyme found in root exudates and root extracts of P-deficient proteoid roots. A, Partially purified sAPase (sAP, 0.04 μg of protein) and cell-free root extracts from +P normal roots (+, N, 0.25 μg of protein), −P normal roots (−, N, 0.15 μg of protein), and −P proteoid roots (−, P, 0.15 μg of protein) stained for acid phosphatase activity on a non-denaturing polyacrylamide gel. B, Partially purified sAPase (sAP, 0.04 μg of protein) and cell-free root extracts from +P normal roots (+, N, 0.15 μg of protein), −P normal roots (−, N, 0.25 μg of protein), and −P proteoid roots (−, P, 0.25 μg of protein) electrophoresed on a non-denaturing gel and then transferred to a membrane, periodate oxidized, and exposed to APase antiserum.
Several compounds were tested as in vitro substrates (at a fixed 1 mm concentration) with the sAPase purified through the (NH4)2SO4 step. Results were calculated on a relative basis to activity obtained by assay of an equivalent amount of enzyme using p-nitrophenol phosphate as a substrate. Although this data cannot be used to infer specificity or catalytic efficiency, results show that the sAPase was able to cleave a phosphate group from ATP (40%), phosphoenolpyruvate (PEP; 34%), NADPH (23%) and Fru-1,6-bisP (23%) with relatively good activity while others (Fru-6-P, Glc-6-P, phytic acid, and Glc-1-P) showed lower activities as potential substrates in comparison to the control.
Initial results from the use of the APase antibodies on immunoblots revealed numerous reactive bands on lanes containing lupin root cell free protein extracts. Since all known APases to date have been determined to be glycoproteins (Schenk et al., 2000), and since it is well known that carbohydrate moieties are highly immunogenic, we hypothesized that many of the bands observed were the result of nonspecific binding of anti-glycan antibodies to plant glycoproteins. To remove this nonspecific cross-reaction, membranes were pretreated with 10 mm metaperiodate to oxidize glycoprotein glycans (Laine and Faye, 1988). Figure 1B shows the result after a native PAGE gel (identical to that run in Fig. 1A) immunoblot following periodate oxidation. The APase antiserum reacted only with isoform 2 in the −P normal and proteoid root extract lanes and with a single band in the purified sAPase lane. The antiserum raised against the sAPase did not recognize the constitutive APase isoform 1, underscoring the distinct nature of these two isoforms.
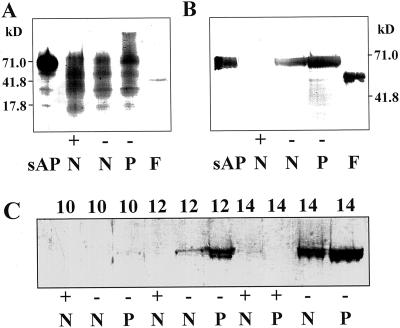
In order to detect the presence of Man and Glc residues generally found on plant proteins, purified sAPase, purified sAPase fusion protein, and normal and proteoid root cell-free extracts from +P and −P plants were electrophoresed on SDS-PAGE Phast gels (Pharmacia, Piscataway, NJ) and blotted to Immobilon P membrane (Millipore, Bedford, MA). Detection of glycan moieties using the periodic acid/Schiff reagent (Strömqvist and Gruffman, 1992) was unsuccessful, whereas the concanavalin A (ConA)-biotin/avidin-alkaline phosphatase system gave clearly positive results on the purified sAPase lanes (Fig. 2A). The fusion protein also appears to be glycosylated, but it is unknown whether the bacterially produced fusion protein is glycosylated in the plant sAPase portion, the T7 phage gene 10 portion, or both. A smear of reactive bands was observed in the lanes containing cell-free extracts, demonstrating that root tissue contains numerous glycoproteins.
Figure 2.
Acid phosphatase secreted from P-deficient proteoid roots is a glycoprotein. A, SDS-PAGE gel blot stained with ConA-biotin to detect glycoproteins in partially purified lupin sAPase (0.08 μg of protein), purified sAPase fusion protein (F, 0.022 μg of protein), and 14- to 16-DAE cell-free extracts from +P normal roots (+, N, 0.25 μg of protein), −P normal roots (−, N, 0.15 μg of protein), and −P proteoid roots (−, P, 0.15 μg of protein). B, SDS-PAGE gel immunoblot using APase antiserum following periodate oxidation showing expression of the APase protein in 14- to 16-DAE cell-free extracts from +P normal root (+, N, 5 μg of protein), −P normal root (−, N, 5 μg of protein), and −P proteoid root (−, P, 5 μg of protein) tissues of 16-DAE white lupin. Lanes sAP (0.30 μg of protein) and F (0.040 μg of protein) contain partially purified lupin sAPase and purified APase fusion protein, respectively. C, SDS-PAGE gel immunoblot using APase antiserum following periodate oxidation showing expression of the APase protein in cell-free extracts from developing roots of lupin. Each lane contains 12.5 μg of protein from normal (N) or proteoid (P) root tissue under +P (+) or −P (−) conditions from 10 to 14 DAE. The numbers at the side of each blot indicate the molecular mass of protein markers in kD.
Electrophoresis of +P and −P cell-free extracts and the purified sAPase protein revealed that the APase activity could be separated into two divergent groups based on their pI. One group, containing two bands of APase activity, had pIs near 5 while the other group, composed of at least six activity bands, migrated to the opposite end of the gradient, near the pH 8 marker (data not shown). The purified sAPase migrated to the pH 5 end of the gel as two bands of activity (identical in migration to the two acidic activity staining bands observed in the −P normal and proteoid root extracts). The +P normal root extract contained only the bands near pH 8, whereas the −P normal and proteoid extract contained both groups in differing quantities. The difference in the degree of glycosylation is a likely explanation for the numerous bands of activity observed on the isoelectric focusing (IEF) gels. An IEF gel was also blotted on Immobilon P membrane, periodate oxidized, and stained with APase antibodies. The serum reacted only with the two activity bands observed at the acidic end of the gel, and therefore no bands were observed for the +P normal root extract (data not shown).
Periodate-treated immunoblots made from SDS-PAGE gels were also probed with sAPase antibodies. Figure 2B shows that the antiserum recognized a single band of apparent molecular mass of 69.6 kD in the purified sAPase lane. The serum also recognized a single band in the APase fusion protein lane (54.8-kD apparent molecular mass), which was used in a final booster for antiserum production. Although no major staining polypeptide bands were observed in the +P normal root lane, both lanes representing −P root tissue showed one reactive band. The lack of a reactive polypeptide in the +P normal root lane is in agreement with the results presented in Figure 1 in which neither enzyme activity nor immunoreactive polypeptide was observed for isoform 2 in the +P normal root extract lanes. Proteoid roots from −P plants had the most intensely staining polypeptide band, correlating with the increased band intensity on native gels stained for enzyme activity (Fig. 1A). An SDS-PAGE gel treated as outlined for Figure 2B was also run to evaluate the appearance of sAPase during lupin root development (Fig. 2C). When cell-free extracts from developing lupin root tissues were electrophoresed and probed with the APase antiserum, immunoreactive polypeptides were observed primarily in the −P tissue in both the normal and proteoid roots at increasing levels from 10 DAE to 14 DAE of development. No increase in immunoreactive polypeptide was observed in +P normal or proteoid root tissue over the time course. This result correlates well with enzyme activity in root extracts as published by Gilbert et al. (1999).
Isolation of a Complete cDNA Encoding sAPase from a White Lupin Proteoid Rootlet cDNA Library
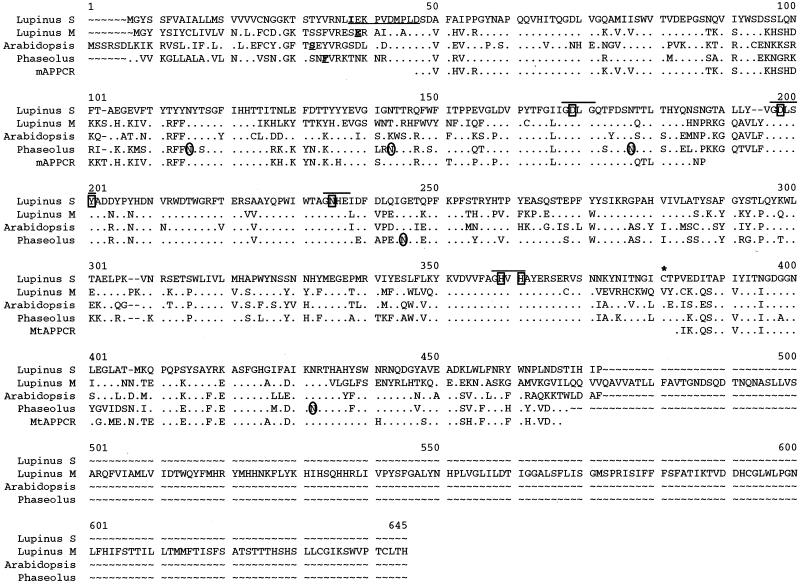
The complete cDNA for the sAPase was obtained by probing a lupin proteoid root cDNA library with a PCR product synthesized from 14-DAE lupin proteoid root first-strand cDNA. The 879-bp PCR product used as the probe was generated using degenerate primers designed by comparison of conserved regions of sequence for several APase cDNAs. The deduced amino acid sequences of these proteins are shown in Figure 3, while the nucleic acid sequence of the white lupin sAPase reported here can be obtained from GenBank as accession no. AF309552. The sAPase cDNA recovered from the library was 68.3%, 70.8%, and 65.9% identical on the protein level to the white lupin, Phaseolus vulgaris, and Arabidopsis APase cDNAs from which the primers were designed. The sAPase cDNA was 1,532 bp in length with an open reading frame of 1,380 bp capable of encoding a 460-amino acid protein. The estimated molecular mass of this protein is 52,473 D with a calculated pI of 5.4. This lupin sAPase cDNA shares several amino acid sequence similarities with the previously isolated cDNAs, including conservation of the position and sequence surrounding metal ligating residues (Klabunde et al., 1995; Schenk et al., 2000), the position of possible sites of N-linked glycosylation (Klabunde et al., 1994), and the location of a conserved Cys residue possibly involved in a disulfide bridge joining the two monomers found in many APase proteins (see Fig. 3; Durmus et al., 1999).
Figure 3.
Comparison of the deduced amino acid sequences of four complete APase proteins and two APase PCR products. The underlined, bold single amino acids indicate the start of the mature white lupin (Lupinus) mAPase (GenBank accession no. AB0233385), Arabidopsis purple acid phosphatase (PAP) (SwissProt accession no. Q38924), and P. vulgaris PAP (GenBank accession no. AJ001270) proteins. The 10 underlined amino acids in the white lupin sAPase sequence indicate the amino acids determined from N-terminal analysis of the purified sAPase protein. Bold lines are above blocks of amino acid residues surrounding the seven metal ligating residues (indicated by a box). The conserved Cys residue (marked by an asterisk) represents a possible site of disulfide bridge between monomers. The circled Asn (N) residues represent glycosylation sites previously determined for the P. vulgaris PAP. Identical residues are identified by dots (●). Dashed lines (∼) indicate gaps introduced in the sequences to maximize similarity. Amino acid position numbers are indicated above the sequence (Lupinus S, lupin sAPase; Lupinus M, lupin mAPase; Arabidopsis, Arabidopsis-secreted PAP; Phaseolus, red kidney bean PAP; mAPPCR, mAPase PCR product; MtAPPCR, Medicago truncatula APase PCR product).
N-terminal sequence analysis of the purified, electroeluted sAPase protein revealed a perfect 10-amino acid match with amino acids 32–41 of the predicted protein sequence of the sAPase cDNA (Fig. 3). This result demonstrates that the sAPase cDNA obtained encodes the secreted form of the enzyme and also defines a 31-amino acid presequence. In addition, PSORT analysis of the sAPase presequence indicates that this protein is directed to the outside of the cell (secreted) with a certainty of 0.82. The predicted molecular mass of the processed protein is 49,210 D. This molecular mass does not take into consideration any mass introduced by glycosylation of the protein and therefore does not coincide with the molecular mass of the purified protein as predicted by SDS-PAGE gels.
Comparison of sAPase mRNA and a Previously Characterized LASAP1 Lupin Membrane APase (mAPase) mRNA Expression in White Lupin
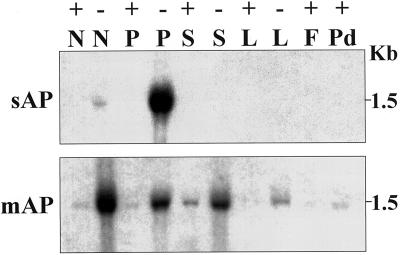
To more fully evaluate APase isoform transcript accumulation, DNA fragments corresponding to sAPase and mAPase (Wasaki et al., 1999) were used to probe RNA blots. The mAPase PCR product used for probing was designed to specifically hybridize to a divergent region of mAPase. The 425-bp product obtained was 97.1% identical to the mAPase from which the primers were made and 79.2% identical to the sAPase cDNA sequence. The deduced amino acid sequence for the mAPase PCR product is shown in Figure 3. As shown in Figure 4 with total RNA loads of 15 μg per lane, both the sAPase and mAPase probes hybridize with a 1.5-kb mRNA, but transcript accumulation for the two was strikingly different. Transcripts for sAPase were highly expressed in −P 14-DAE proteoid roots. Expression of sAPase was also detected in normal roots of −P plants at 14 DAE, but steady-state amounts were much lower as compared to that seen in proteoid roots. When total RNA load per lane was increased to 30 μg (data not shown), transcript accumulation for the sAPase was observed to steadily increase over time from 10 DAE to 14 DAE in both P-deficient normal and proteoid tissues. Little or no change in transcript accumulation was observed in P-sufficient tissue for sAPase transcript over time through 14 DAE. In contrast, transcripts for mAPase were detected in greatest amounts in −P normal roots at 14 DAE, accompanied by expression at reduced levels in leaves, stems, and proteoid roots of −P plants. Detectable amounts of mAPase transcripts were also found in +P treatments. This differential pattern of transcript accumulation further reflects the distinct nature of these APase genes.
Figure 4.
RNA gel-blot analysis of the expression of two lupin APases in various P-sufficient and P-deficient tissues of lupin. Each lane contains 15 μg of total RNA from normal root (N), proteoid root (P), stem (S), leaf (L), flower (F), or pod (Pd) tissue under +P (+) or −P (−) conditions. The blots were hybridized with inserts specific for either sAPase (sAP) or mAPase (mAP). The numbers at the right indicate the size of the hybridizing band from each blot in kilobases.
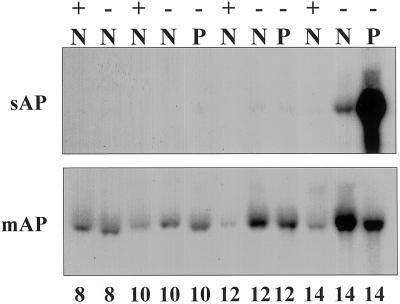
Because sAPase cDNA encodes a secreted form of acid phosphatase from proteoid roots, we thought it important to evaluate the expression of sAPase transcripts during development of normal and proteoid roots under P-sufficient (+) or P-deficient (−) conditions. Normal and proteoid roots were harvested from +P or −P plants between 8 and 14 DAE (Fig. 5). Expression of sAPase was dramatically stimulated in −P proteoid roots at 14 DAE, with much less occurring in normal roots of −P plants. At 12 DAE, sAPase transcripts were detectable in both normal and proteoid roots of −P-grown plants. In contrast, mAPase transcripts were detected in both normal and proteoid roots at each time point, with greatest expression occurring in normal roots of −P plants at 14 DAE.
Figure 5.
RNA gel-blot analysis of the expression of two lupin APases in developing P-sufficient and P-deficient root tissues of lupin. Each lane contains 15 μg of total RNA from normal (N) or proteoid (P) root tissue under +P (+) or −P (−) conditions. Samples were collected from 8 to 14 DAE. The blots were hybridized with inserts encoding either the sAPase (sAP) or mAPase (mAP). Note that emerged proteoid roots are not present on plants 8 DAE, so whole roots are collected.
Phosphate Deficiency and Al Treatment Increase Expression of the sAPase Transcript
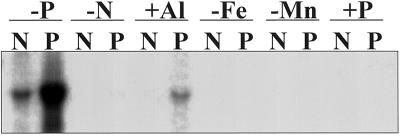
The effect of various nutrient stresses on sAPase expression was determined by RNA gel-blot analysis of stressed root tissues as shown in Figure 6. Previous studies have shown that macro- and micronutrient stress in white lupin alters the proteoid root mass when expressed as a percentage of total root mass, as well as affecting total shoot and root growth (Johnson et al., 1994). All stress treatments were carried out under P-sufficient conditions. Proteoid roots typically represent approximately 10% of the total root mass on P-sufficient white lupin plants (Johnson et al., 1996). Transcript for sAPase was detected in proteoid roots starved for N (at low levels) and in proteoid roots (at higher levels) treated with Al. The sAPase transcript was not detected in either root tissue type in the −Fe, −Mn, or +P treatments. The sAPase transcript showed expression levels for −P normal and proteoid root tissue comparable to that seen in Figure 5.
Figure 6.
RNA gel-blot analysis of the expression of sAPase in lupin normal (N) or proteoid (P) root tissue under various nutrient stresses. Each lane contains 15 μg of total RNA isolated from P-deficient (−P), N-deficient (−N), Fe-deficient (−Fe), or Mn-deficient (−Mn) root tissue and Al-treated (+Al) or P-sufficient (+P) root tissue. The N-, Fe-, Mn-, and Al-treated plants were grown under P-sufficient conditions.
Genomic Organization of Two APase Genes and Isolation of the sAPase Promoter
DNA gel-blot analysis was used to determine gene copy number for both sAPase and mAPase (data not shown, available upon request). Under highly stringent wash conditions, a single hybridizing fragment was observed in all lanes for sAPase. A replicate blot was probed with the mAPase fragment. All hybridizing bands (a single band in each lane) for mAPase differed from those observed for sAPase. Additional bands were observed on a blot probed with the mAPase fragment when it was washed under low stringency conditions. Some of the additional bands on the blot washed under low stringency conditions were identical to those bands observed on the sAPase blot washed at high stringency. The band separation pattern further confirms the distinct nature of sAPase and mAPase at the gene level.
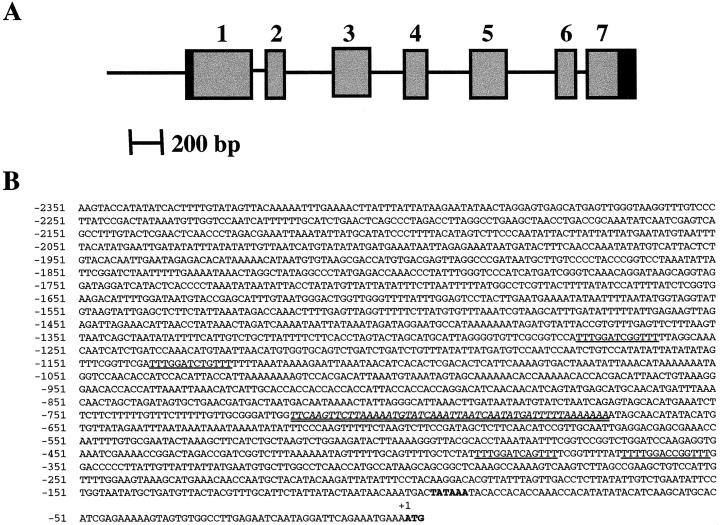
For future studies to delimit cis-elements and trans-acting factors contributing to plant adaptation to low P environmental stress, we thought it important to isolate the gene encoding the sAPase enhanced in −P lupin proteoid roots. Several positive clones were obtained from a partial genomic library and one, which hybridized with the 5′-most portion of the cDNA, was found to contain sequences corresponding to the full-length sAPase cDNA. A 5.3-kb region of this genomic clone was sequenced and found to include the entire coding region and approximately 2.5 kb upstream of the ATG start site. The sequence of transcribed region of the gene can be found in GenBank accession no. AF317218, and the intron-exon structure of white lupin sAPase is shown in Figure 7A. Seven exons ranging from 119 bp to 369 bp in length are interrupted by six introns ranging from 69 bp to 307 bp. All intron-exon junctions are bordered by the expected GT-AG consensus sequence. The sequence of the putative promoter 2.5 kb 5′ to the ATG is shown in Figure 7B. A putative TATA box sequence was located between −96 and −91 bp upstream of the ATG start site. A homology search in GenBank did not reveal significant overall homology with any promoter registered; however, bestfit analysis of the lupin promoter region with the Arabidopsis APase gene (located on chromosome II; Lin et al., 1999; Haran et al., 2000) did reveal a 50-bp region within the lupin APase promoter with 72% sequence identity to the Arabidopsis APase promoter. Both APase genes also share identical intron-exon junctions, though the intron sizes differ. Computer analysis of the lupin APase promoter sequence identified four imperfect repeats with the consensus sequence 5′-TTTGGAPyCNGTTT-3′. Functional analysis of the APase promoter is now being carried out to determine whether these repeats contribute to the expression pattern under −P conditions.
Figure 7.
A, Diagrammatic representation of the structure of the sAPase gene from lupin. Exons are indicated by boxed regions, while introns and the 5′ non-transcribed region are represented by lines. The blackened portion of exons 1 and 7 correspond to the untranslated sequences. B, Sequence of the 5′-upstream region of the sAPase gene. The translational start codon and the putative TATA box are shown in bold. The repeats with consensus core sequence 5′-TTTGGAPyCNGTTT-3′ are underlined. The 50-bp region with 72% sequence identity to the Arabidopsis APase promoter is indicated in italics and double underlined.
DISCUSSION
Plants subjected to a P-deficient environment exhibit a number of biochemical, developmental, and molecular responses (Schachtman et al., 1998; Raghothama, 1999). Here, we extend the understanding of plant adaptation to P deficiency by defining the interrelationship between proteoid root development in white lupin and secretion of APase protein into the rhizosphere. Release of sAPase into the rhizosphere of proteoid roots from P-deficient plants involves enhanced synthesis of sAPase protein and mRNA. Release of sAPase into the rhizosphere is accompanied by cleavage of a 31-amino acid precursor. The white lupin sAPase is a glycoprotein with broad substrate specificity. Moreover, the promoter of the white lupin sAPase gene has a 50-bp region strikingly similar to the Arabidopsis P-responsive APase promoter and can direct transient reporter gene expression to proteoid roots.
Proof that the APase protein purified in this study represents the secreted form of the enzyme and, furthermore, that the cDNA isolated encodes the secreted form of the protein comes from several lines of evidence. Nondenaturing PAGE showed that the purified enzyme comigrated with isoform 2, previously identified by Gilbert et al. (1999) as an APase isoform induced under P-deficient conditions in proteoid roots. N-terminal sequence analysis of the purified sAPase protein resulted in a perfect match of 10 amino acids at positions 32 to 42 in the deduced amino acid sequence of the isolated cDNA. This match defined a 31-amino acid presequence for which PSORT analysis predicted a high probability that the deduced protein was secreted outside of the cell. The deduced protein had a predicted pI of 5.4, whereas the purified sAPase migrated to the pH 5 area on an IEF gel, clearly separated from other forms of APase, which migrated to the opposite (basic, pH 8–9) end of the pH gradient. Transcripts for the sAPase were detected in greatest amounts in P-deficient proteoid root tissue at 14 DAE, accumulation of transcript for this message being observed as early as 10 DAE in −P tissues. Little or no change in sAPase transcript accumulation was observed in +P tissue over time during early plant growth and development.
The sAPase cDNA reported here differs significantly from a white lupin LASAP1 APase cDNA reported previously by Wasaki et al. (1999). Results from analysis of the deduced protein, including PSORT analysis of the proposed 31-amino acid presequence, indicate that LASAP1 likely represents a membrane-bound form of APase found in both root and shoot tissue, and therefore does not encode the APase isoform purified by their colleagues (Ozawa et al., 1995). Using this mAPase sequence and sequence information from an expressed sequence tag isolated from our lupin cDNA library to design primers, a PCR product was generated that allowed us to distinguish between transcript expression for the two APase forms. Results of the RNA and DNA gel blots confirmed the distinct nature of these two isoforms. Whereas the sAPase transcripts are observed mainly in P-deficient proteoid roots, the expression of mAPase was detected at its highest level in P-deficient normal roots and at lower but significant levels in leaves, stems, and proteoid roots of P-deficient plants. This is in agreement with previous results for the mAPase in as far as results were presented on the basis of P regime in roots or shoots (Wasaki et al., 1999) but results here further localize the increased expression of the mAPase transcript in P-deficient roots to normal root tissue, and not proteoid root tissue where the sAPase is predominantly expressed. We calculated the size of the hybridizing mAPase RNA on gel blots presented here to be 1.5 kb, whereas the previously isolated mAPase cDNA was reported to be 2.2 kb (Wasaki et al., 1999). DNA gel-blot analysis, using the mAPase PCR product as a probe, agrees with data from Wasaki et al. (1999) for the size estimate of the hybridizing HindIII band. While our blots show a faint hybridizing EcoRI band near the 9.4-kb marker in agreement with the Tadano group, our blots also displayed a strong band just below the 4.3-kb marker not observed on their DNA gel blots. The unusual 3′-terminal region contained on this cDNA has not been reported for any other plant, animal, or fungal APase sequenced to date (Schenk et al., 2000). One possibility is that our mAPase PCR fragment represents another APase form that is closely related but yet distinct from the previously reported LASAP1 mAPase form. The calculated pI for the LAPSAP1 mAPase protein deduced from the cDNA sequence published previously was 8.0, placing it at the opposite end of the gel from the −P proteoid root sAPase isoform.
The apparent molecular mass of the purified sAPase polypeptide, as calculated from SDS-PAGE gels, was approximately 70 kD, in close agreement with the molecular mass reported by Ozawa et al. (1995) for a lupin-secreted APase. The estimated molecular mass of the deduced protein from the cDNA reported here was 49.2 kD. The difference in predicted versus apparent mass is very likely due to the presence of carbohydrate moieties on the protein, shown to be present by ConA-biotin staining, which would increase the mass of the protein and often has been found to affect the mobility of a protein in gel systems (Hames, 1981). Variation in the number of carbohydrate groups on the protein also likely accounts for the numerous bands of APase activity observed on IEF gels. The carbohydrate content of the protein also apparently affected the overall quality of the antiserum produced against the protein, resulting in a mixture of antibodies produced against both the glycan and peptide epitopes of the protein, a problem noted previously for serum raised against APases and other glycoproteins (Duff et al., 1994; Cashikar et al., 1997; Wasaki et al., 1999). When the glycan epitopes were oxidized via periodate, the antiserum recognized only APase isoform 2 in cell-free extracts and the purified form of the sAPase under both nondenaturing and denaturing conditions. In IEF gels, only those forms of the enzyme that migrated to the acidic end of the gel and, again, the purified sAPase were recognized by the antiserum. These results indicate that other forms of APase found in lupin roots and shoots, such as the membrane form of the enzyme, are not recognized by the antiserum produced by injection of the sAPase protein. Others have noted either cross-reactivity with some but not all isoforms of APase in one plant species or immunological cross-reactivity of APases between plant species (Duff et al., 1994). Immunological distinction of protein isoforms has been noted previously for other proteins (Farnham et al., 1990; Miller et al., 1998).
White lupin P deficiency induced sAPase, similar to other APases, in that it can hydrolyze P from a number of substrates. Synthesis and exudation of an APase that can cleave P from numerous substrates is seemingly an efficient adaptation to P deficiency. In planta, the enzyme could release P from several biologically active phosphate-containing compounds as a mechanism to recycle internal P as demonstrated for PEP APase (Duff et al., 1994). Likewise, exudation into the rhizosphere of proteoid roots would provide enhanced access to organic phosphate esters over a much wider soil area than exudation from normal roots. Although the relative importance of P acquisition from soil organic matter is not well established, the fact that up to 38% of the total soil organic P in some soils is comprised of phytate offers a large pool of potentially available P (Hayes et al., 1999). Even with a significantly reduced efficiency for cleaving organic P from phytate, the copious release of sAPase from P-deficient proteoid roots could release appreciable P in soils with high phytate concentrations, providing additional sources for plant growth.
Due to the scarcity of characterized APase cDNAs and genes, progress in understanding the molecular events that regulate APase activity has been slow. In this report we prove that white lupin sAPase expression is directly related to enhanced accumulation of sAPase mRNA and protein in response to P stress. Moreover, in planta distribution of protein and mRNA is fairly specific for proteoid roots of P-stressed plants. These data suggest that a signal or signals produced in P-stressed proteoid roots activates expression of the sAPase gene. In isolating the sAPase gene and the 5′-upstream putative promoter region of sAPase, we have developed the tools to address this hypothesis. It is noteworthy that within the +1- to −751-bp region upstream of the ATG of white lupin APase lies a 50-bp region that is 72% identical to that found in the promoter of an Arabidopsis mAPase gene (Lin et al., 1999; Haran et al., 2000). This Arabidopsis promoter can direct enhanced gene expression under P-stressed conditions. The similarity between white lupin P stress-induced sAPase and the Arabidopsis mAPase is further evidenced by the fact that the intron-exon structure of these genes is identical. A second Arabidopsis APase gene with an open reading frame of 338 amino acids interrupted by two introns representing a mammalian type 5 APase was also reported recently (del Pozo et al., 1999). The promoter of the type 5 APase is also responsive to ABA and salt stress as well as phosphate starvation (del Pozo et al., 1999). The cDNA for this APase has a leader sequence of 31 amino acids. Our analysis of the 31-amino acid presequence by PSORT of this type 5 APase indicated that it directs the protein outside of the cells; however, del Pozo et al. (1999) found no evidence of the protein in the apoplastic fluid of root tissue. Ongoing work from the M. truncatula genome project has revealed an APase that appears to be induced in P-starved roots (C.P. Vance, unpublished data).
The concept of a phosphate stress-induced regulon existing in plants, as has been shown to occur in bacteria and yeast systems (Goldstein, 1992; Delhaize and Randall, 1995; Malboobi and Lefebvre, 1995), is an especially engaging possibility in white lupin. In this system, not only is the exudation of large amounts of sAPase from the root system controlled by the P status of the plant but, additionally, the plants respond to P stress conditions by initiating a new root type, namely proteoid roots. Therefore, unlike the tissue culture systems reported previously that secrete an sAPase (Goldstein, 1992; LeBansky et al., 1992; Duff et al., 1994; Goldstein et al., 1988), P-deficiency in white lupin gives rise to coordinate regulation of several genes that direct the initiation, development, and subsequent unique functioning of proteoid roots, seemingly with the sole purpose of P acquisition. With regards to P acquisition, it is probable that proteoid roots also secrete other proteins besides APases which, in conjunction with the sAPase, aid in P mobilization for plant nutrition. S-like RNases have been previously implicated as having a possible role in mobilizing P from sources of RNA in the rhizosphere (Nürnberger et al., 1990; Goldstein, 1992; Dodds et al., 1996). Several as yet unidentified proteins have been found to be synthesized under conditions of P stress as noted by several authors (Goldstein, 1992; Malboobi and Lefebvre, 1995; C.P. Vance, unpublished data), some of which appear to be intracellular in location while others are exuded into the surrounding rhizosphere. Other phosphate stress-induced proteins hypothesized include phosphate transporters, protein phosphatases, PEP carboxylase (Goldstein, 1992; Malboobi and Lefebvre, 1995; Johnson et al., 1996; Neumann et al., 1999), and cytosolic malate dehydrogenase (C.P. Vance, unpublished data).
MATERIALS AND METHODS
Plant Materials
White lupin (Lupinus albus L. var. Ultra) was grown in a growth chamber and watered with the appropriate nutrient solution as previously described (Johnson et al., 1994; Gilbert et al., 1999). Nutrient solutions differed only in P concentrations (Johnson et al., 1994; Gilbert et al., 1999), except during the stress experiment in which the solutions designated −Fe, −Mn, and −N were mixed as described by Johnson et al. (1994). The Al stress treatment consisted of the addition of AlK(SO4)2 at a concentration of 450 μm to a nutrient solution that was used to water the plants every other day. A +P nutrient solution was sprayed onto the leaves at the same time the plants were watered in order to supply adequate P to the plants and to avoid precipitation of Al phosphate in the nutrient solution.
Enzyme Purification
The spectrophotometric assay of APase activity using p-nitrophenol phosphate as the substrate was carried out as previously described (Gilbert et al., 1999) to monitor the enzyme throughout the purification. Proteoid root sections were harvested from 14-DAE lupin plants (approximately 130 plants) grown in the absence of P and placed immediately into a large beaker containing 300 mL of room temperature 50 mm maleate buffer (pH 5.5) containing 2% (w/v) Suc, 1 mm phenylmethylsulfonyl fluoride, and 10 μm antipain. Following harvest of all of the proteoid sections (1.5 h), the plant material was placed under vacuum for 5 min and then roots were allowed to exude for 1 h at room temperature. The root sections were then removed and placed in 300 mL of fresh maleate buffer and allowed to exude for an additional 1 h at room temperature. The supernatants were combined and centrifuged at 10,000 rpm for 20 min to remove any remaining root segments or sand debris. The clarified supernatant was fractionated at 4°C with solid (NH4)2SO4. The fraction precipitating between 45% and 80% was collected after overnight incubation at 4°C by centrifugation at 12,000 rpm for 30 min. The pellet was dissolved in a minimum volume of 50 mm maleate buffer (pH 5.5) containing 2% (w/v) Suc and loaded onto four 1.5-mm-thick non-denaturing 10% polyacrylamide gels (Ornstein, 1964) and run at 32 mA constant current for 4 to 5 h. A 1-cm-wide lengthwise section of each gel was cut away for in vivo activity staining as described previously (Gilbert et al., 1999) to locate the major staining band of APase activity. The activity-stained section was realigned with the unstained portion of each gel (stored at 4°C during staining), and the gel area corresponding to the activity was excised. The gel slices were placed in dialysis tubing (30,000 molecular weight cutoff) containing a minimal amount of buffer (25 mm Tris, 190 mm Gly, pH 8.3), and the gel protein was electroeluted at 50 V constant voltage into the tubing buffer at 4°C overnight. The electroelution was carried out for three consecutive nights to assure removal of all the APase protein. Each morning, the elution buffer in the tubing was exchanged for fresh buffer following reversal of polarity at 100 V for 5 min. Bradford protein assays (Bio-Rad, Hercules, CA) were performed on each of the collected elution aliquots to determine the yield of enzyme protein. Purity was evaluated by electrophoresing the electroeluted protein on both SDS-PAGE and non-denaturing PAGE systems and staining with Coomassie blue or silver. Subunit molecular mass was estimated by running prestained molecular mass standards (Bio-Rad) and electroeluted protein on an SDS-PAGE gel and staining with silver. A portion of the electroeluted protein sample was submitted for N-terminal sequence analysis to the Microchemical Facility of the Institute of Human Genetics (University of Minnesota, Minneapolis). The enzyme protein was concentrated for injection into rabbits by placing dialysis tubing containing the protein on a bed of solid Suc.
Substrate Specificity
Secreted APase protein purified through the (NH4)2SO4 step was used to test substrate specificity. Enzyme activity was quantitated by measurement of inorganic phosphate released as described by Olczak et al. (1997). Substrates were dissolved in 0.1 m sodium acetate buffer, pH 5.0 and used in the 1-mL assays at a final concentration of 1 mm. The enzyme reaction was allowed to run for 30 min at 37°C before being stopped by addition of SDS. All assays were linear with respect to time and enzyme concentration. A phosphate standard curve was generated using 25 to 150 μL of a 1 mm KH2PO4 standard solution. Parallel assays using p-nitrophenol phosphate as a substrate were run using the same quantity of enzyme, and the amount of p-nitrophenol released was measured at 410 nm.
Fusion Protein Production and Purification
The pGEMEX T7 expression vector system (Promega, Madison, WI) was used for high-level expression of the N-terminal portion of the protein encoded by the sAPase cDNA clone. A 390-bp (bp 34–416 of the sAPase cDNA) PCR product was generated using the 5′ primer 5′-GGGGAATTCATGGGTTATAGTAGTTTTTGT-3′ and the 3′ primer 5′-GAGGGATCCTATGTAGTGTCAAACTCCAA-3′. Following transformation into JM109 (DE 3) cells, colonies were selected and induced to produce the fusion protein by the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside as specified by the manufacturer. Cells from cultures producing the predicted 40-kD product of the T7 gene 10-APase fusion were resuspended after centrifugation in phosphate-buffered saline (10 mm K2HPO4, 150 mm NaCl, pH 7.2), and stock lysozyme (50 mg mL−1) was added to 110 μg mL−1 cells. The tubes were placed at 30°C for 50 min and then recentrifuged. The pellet was resuspended in 1× SDS protein sample buffer (Maizel, 1971), then boiled for 10 min and loaded onto a 10% SDS-PAGE gel. A 1-cm-wide lengthwise section of the gel was cut away for staining with Coomassie blue to locate the fusion product. The stained section was realigned with the unstained portion of the gel (stored at 4°C during staining), and the gel area corresponding to the activity band was excised.
Antiserum Production and Immunotitration of APase Activity
Polyclonal antiserum was produced against the secreted form of APase by injection of rabbits (New Zealand White) with a mixture of gel-purified, secreted APase protein and AP fusion protein. The first injection contained approximately 350 μg of secreted APase protein in 1.6 mL mixed with 1.45 mL of incomplete and 0.15 mL of complete Freund's adjuvant. Three additional injections (all mixed 1:1 with incomplete adjuvant) were made containing 200 and 150 μg of secreted protein, with the final injection containing 150 μg of the APase fusion protein (Miller et al., 1998). Blood serum was collected and concentrated as previously described (Vance et al., 1985). Immunotitration of APase activity was carried out as described (Miller et al., 1998).
Protein Electrophoresis and Immunoblotting
Total soluble proteins from 14- to 16-DAE lupin normal and proteoid root tissues were prepared as previously described (Gilbert et al., 1999). Proteins were electrophoresed on 10% SDS-PAGE minigels and then electrophoretically transferred to nitrocellulose for exposure to APase antiserum (Vance et al., 1985). Proteins were electrophoresed on native 10% to 15% gradient or IEF (pH 3–9) Phast gels and either stained for enzyme activity (Gilbert et al., 1999) or transferred to Immobilon P membranes as described previously (Gronwald and Plaisance, 1998). Prior to exposure to APase antiserum, immunoblots were incubated for 2 h at room temperature (in the dark) in 0.1 m sodium acetate buffer, pH 4.5, containing 10 mm sodium metaperiodate followed by a 30-min room temperature incubation in 50 mm sodium borohydride in phosphate-buffered saline (Laine and Faye, 1988). Periodate-treated native, IEF, and SDS membranes were blocked for 3 to 6 h using a high-salt buffer system described by Gronwald and Plaisance (1998). The blots were incubated overnight with the APase antibody (1:500 dilution) and then developed using the goat anti-rabbit IgG horseradish peroxidase conjugate system (Vance et al., 1985). A ConA-biotin/avidin-alkaline phosphatase detection system was used on non-periodate oxidized Immobilon P membranes blotted from denaturing Phast gels (10%–15% gradient) to detect protein glycosylation (Gronwald and Plaisance, 1998).
cDNA Library Construction and Screening
A cDNA synthesis kit (Stratagene, La Jolla, CA) was used to construct an oligo(dt)-primed white lupin proteoid root library in the excision vector lambda ZAPII. Twelve- (4 μg) and 14-DAE (3.1 μg) proteoid rootlet poly(A+) RNA were combined and treated with methylmercury hydroxide and used for construction of the library, which yielded 1.6 × 106 original transformants. The amplified library was screened for a secreted APase isoform with a PCR probe generated by reverse transcriptase-PCR using degenerate primers designed based on known APase sequences. The 5′ primer was 5′-CTCARCAGGTTCATRTAACRCAAGG-3′ and the 3′ primer was 5′-GNCCNGCRAANACAACRTCAAC-3′. Reverse transcription was carried out on total lupin proteoid root RNA (14 DAE) using Superscript II (Gibco-BRL, Gaithersburg, MD). A band of approximately the expected size was gel purified from the PCR mixture and ligated into the pGEM-T vector. The insert was sequenced and then radiolabeled and used to probe the lupin proteoid root cDNA library. A full-length APase cDNA was subsequently isolated (named sAPase) and sequenced in its entirety. The PSORT program was used for the prediction of protein localization sites in cells (Nakai and Kanehisa, 1992).
PCR of Membrane Acid Phosphatase
Primers were designed for PCR based on a known white lupin membrane APase (LASAP1; Wasaki et al., 1999) and a lupin membrane APase expressed sequence tag (isolated from the cDNA library described here) to generate a DNA fragment representing a lupin membrane APase. The sequence of the 5′ primer was 5′-GATAGCGATGTATTTCATGTCC-3′ and the sequence of the 3′ primer was 5′-CTTGGGTTATGTTGATAGTGAG-3′. PCR was performed using a phenol-treated aliquot of the 12- to 14-DAE lupin proteoid rootlet cDNA library in the presence of 5% (v/v) methylsulfoxide. A band of the expected size (425 bp) was obtained and purified using a Qiaquick PCR kit (Qiagen, Valencia, CA) and then ligated into a pGEM-T vector (Promega). The insert was sequenced in its entirety. A fragment for radiolabeling was generated by plasmid restriction using NotI and NcoI restriction enzymes.
RNA and DNA Gel-Blot Analysis
Total RNA was isolated from frozen lupin root, stem, leaf, flower, and pod tissue by methods described elsewhere (DeVries et al., 1982). Total RNA was electrophoresed and transferred as described by Johnson et al. (1996). RNA blots were probed and washed under high stringency conditions using the formamide protocol as per the manufacturer's instructions (Bio-Rad). Radioactivity on RNA blots was quantitated using an AMBIS radioanalytic imaging system (Scanalytics, Billerica, MA). Equal lane loading was verified by probing blots with a 28S rRNA subunit from Phaseolus vulgaris. Genomic DNA was extracted from young lupin leaves as described by Junghans and Metzlaff (1990). Ten micrograms of genomic DNA was digested with EcoRI, HindIII, XbaI, or PstI and fractionated on a 0.8% (w/v) agarose gel. DNA blots were generated by transfer of the DNA to Immobilon NY+ membrane (Millipore) and then hybridized at 68°C and washed under high stringency conditions (68°C, 0.1× SSC, 0.1% SDS).
Construction of a Partial Genomic Library and Isolation of an Acid Phosphatase Gene
Ten micrograms of genomic DNA from white lupin was restricted with EcoRI, separated on a 0.6% (w/v) agarose gel, and blotted to nylon membrane. The blot was probed with the 32P-labeled sAPase cDNA clone from lupin, and a 15-kb genomic fragment was recognized by the probe. A second larger digest (50 μg) was then carried out with EcoRI, and the fragments were run on several lanes of an agarose gel. DNA fragments in the area of 15 kb were cut from the gel, purified by phenol:CHCl3:isoamyl alcohol extraction followed by ethanol precipitation, and then cloned into a lambda DASH II EcoRI cut vector (Stratagene). The partial library contained 500,000 original transformants. It was subsequently amplified and 500,000 plaque-forming units were plated and probed using the radiolabeled sAPase cDNA described above. Following three rounds of plating, one positive clone was identified. Lambda DNA was isolated, and the entire insert was removed with EcoRI and subcloned into pBluescript KS+ (Stratagene). Additional subcloning from pBluescript was undertaken to aid in sequencing of the area of interest. A total of 5.3 kb was sequenced from this genomic clone, which included the entire coding region and aproximately 2.5 kb of the promoter region.
Footnotes
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. USDA/98–35100–6098) and by the U.S. Department of Agriculture-Agricultural Research Service (grant no. CRIS 3640–21000–014–00D). This is a joint contribution of the United States Department of Agriculture-Agricultural Research Service and the Minnesota Agricultural Experiment Station Scientific Journal Series.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010097.
LITERATURE CITED
- Abelson PH. A potential phosphate crisis. Science. 1999;283:2015. doi: 10.1126/science.283.5410.2015. [DOI] [PubMed] [Google Scholar]
- Bieleski RL. Phosphate pools, phosphate transport and phosphate availability. Annu Rev Plant Physiol. 1973;24:225–252. [Google Scholar]
- Cashikar AG, Kamaresan R, Madhusudhana Rao N. Biochemical characterization and subcellular localization of the red kidney bean purple acid phosphatase. Plant Physiol. 1997;114:907–915. doi: 10.1104/pp.114.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1995;107:207–213. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Layva A, de la Peña A, Aragoncilla C, Paz-Ares J. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress conditions. Plant J. 1999;19:579–589. doi: 10.1046/j.1365-313x.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- DeVries FC, Springer J, Wessels JGH. Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and green pea seedlings. Planta. 1982;156:129–135. doi: 10.1007/BF00395427. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Clarke AE, Newbigin E. Molecular characterization of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Mol Biol. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant. 1994;90:791–800. [Google Scholar]
- Durmus A, Eicken C, Spener F, Krebs B. Cloning and comparative protein modeling of two purple acid phosphatase isozymes from sweet potatoes (Ipomoea batatas) Biochim Biophys Acta. 1999;1434:202–209. doi: 10.1016/s0167-4838(99)00176-4. [DOI] [PubMed] [Google Scholar]
- Farnham MW, Miller SS, Griffith SM, Vance CP. Aspartate aminotransferase in alfalfa root nodules: II. Immunological distinction between two forms of the enzyme. Plant Physiol. 1990;93:603–610. doi: 10.1104/pp.93.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. Acid phosphatase activity in phosphorus-deficient white lupin roots. Plant Cell Environ. 1999;22:801–810. [Google Scholar]
- Goldstein AH. Phosphate starvation inducible enzymes and proteins in higher plants. In: Wray JL, editor. Society for Experimental Biology Seminar Series 49: Inducible Plant Proteins. Cambridge, UK: Cambridge University Press; 1992. pp. 25–44. [Google Scholar]
- Goldstein AH, Baertlein DA, McDaniel RG. Phosphate starvation inducible metabolism in Lycopersicon esculentum. I. Excretion of acid phosphatase by tomato plants and suspension-cultured cells. Plant Physiol. 1988;87:711–715. doi: 10.1104/pp.87.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald JW, Plaisance KL. Isolation and characterization of glutathione s-transferase isozymes in sorghum. Plant Physiol. 1998;117:877–892. doi: 10.1104/pp.117.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames BD. An introduction to polyacrylamide gel electrophoresis. In: Hames BD, Rickwood D, editors. Gel Electrophoresis of Proteins: A Practical Approach. Oxford: IRL Press; 1981. pp. 1–91. [Google Scholar]
- Haran S, Logendra S, Saskar M, Bratanova M, Raskin I. Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiol. 2000;124:615–626. doi: 10.1104/pp.124.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Richardson AE, Simpson RJ. Phytase and acid phosphatase activities in extracts from roots of temperate pasture grasses and legume seedlings. Aust J Plant Physiol. 1999;26:801–809. [Google Scholar]
- Johnson JF, Allan DL, Vance CP. Phosphorus stressed-induced proteoid roots show altered metabolism in Lupinus albus. Plant Physiol. 1994;104:657–665. doi: 10.1104/pp.104.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JF, Vance CP, Allan DL. Phosphorus deficiency in Lupinus albus: altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase. Plant Physiol. 1996;112:31–41. doi: 10.1104/pp.112.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans H, Metzlaff M. A simple and rapid method for the preparation of total plant DNA. Biotechniques. 1990;8:176. [PubMed] [Google Scholar]
- Klabunde T, Stahl B, Suerbaum H, Hahner S, Karas M, Hillenkamp F, Krebs B, Witzel H. The amino acid sequence of the red kidney bean Fe(III)-Zn(II) purple acid phosphatase: determination of the amino acid sequence by a combination of matrix-assisted laser desorption/ionization mass spectrometry and automated Edman sequencing. J Biochem. 1994;226:369–375. doi: 10.1111/j.1432-1033.1994.tb20061.x. [DOI] [PubMed] [Google Scholar]
- Klabunde T, Sträter N, Krebs B, Witzel H. Structural relationship between the mammalian Fe(III)-Fe(II) and the Fe(II-I)-Zn(II) plant purple acid phosphatases. FEBS Lett. 1995;367:56–60. doi: 10.1016/0014-5793(95)00536-i. [DOI] [PubMed] [Google Scholar]
- Laine AC, Faye L. Significant immunological cross-reactivity of plant glycoproteins. Electrophoresis. 1988;9:841–844. doi: 10.1002/elps.1150091210. [DOI] [PubMed] [Google Scholar]
- LeBansky BR, McKnight TD, Griffing LR. Purification and characterization of a secreted purple phosphatase from soybean suspension cultures. Plant Physiol. 1992;99:391–395. doi: 10.1104/pp.99.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tadano T. Comparison of characteristics of acid phosphatases secreted from roots of lupin and tomato. Soil Sci Plant Nutr. 1996;42:753–763. [Google Scholar]
- Lin X, Kaul S, Rounsley SD, Shea TP, Benito M-I, Town CD, Fujii CY, Mason TM, Bowman CL, Barstead ME. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Maizel JV. Polyacrylamide gel electrophoresis of viral proteins. In: Maramorosch K, Koprowski H, editors. Methods in Virology. V. New York: Academic Press; 1971. pp. 179–224. [Google Scholar]
- Malboobi MA, Lefebvre DD. Isolation of cDNA clones of genes with altered expression levels in phosphate-starved Brassica nigra suspension cells. Plant Mol Biol. 1995;28:859–870. doi: 10.1007/BF00042071. [DOI] [PubMed] [Google Scholar]
- Miller SS, Driscoll BT, Gregerson RG, Gantt JS, Vance CP. Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J. 1998;15:173–184. doi: 10.1046/j.1365-313x.1998.00192.x. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Massonneau A, Martinoia E, Romheld V. Physiological adaptations to phosphorus deficiency during proteoid root development in white lupin. Planta. 1999;208:373–382. [Google Scholar]
- Nürnberger T, Abel S, Jost W, Glund K. Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol. 1990;92:970–976. doi: 10.1104/pp.92.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olczak M, Watorek W, Morawiecka B. Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds. Biochim Biophys Acta. 1997;1341:14–25. doi: 10.1016/s0167-4838(97)00055-1. [DOI] [PubMed] [Google Scholar]
- Ornstein L. Disc gel electrophoresis: II. Method and application to human serum protein. Ann NY Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Osaki M, Matsui H, Honma M, Tadano T. Purification and properties of acid phosphatase secreted from lupin roots under phosphorus-deficiency conditions. Soil Sci Plant Nutr. 1995;41:461–469. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Runge-Metzger A. Closing the cycle: obstacles to efficient P management for improved global food security. In: Tiessen H, editor. Phosphorus in the Global Environment. Chichester, UK: John Wiley; 1995. pp. 27–42. [Google Scholar]
- Schachtman D, Reid RJ, Ayling SM. Phosphorus uptake by plants from soil to cell. Plant Physiol. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, deJersey J. Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene. 2000;250:117–125. doi: 10.1016/s0378-1119(00)00186-4. [DOI] [PubMed] [Google Scholar]
- Strömqvist M, Gruffman H. Periodic acid/Schiff staining of glycoproteins immobilized on a blotting matrix. Biotechniques. 1992;13:744–746. [PubMed] [Google Scholar]
- Tarafdar JC, Claasen N. Organic phosphorus compounds as a phosphorus source for higher plants through the activity of phosphatases produced by plant roots and microorganisms. Biol Fertil Soils. 1988;5:3308–3312. [Google Scholar]
- Vance CP, Boylan KLM, Stade S, Somers DA. Nodule specific proteins in alfalfa (Medicago sativa L.) Symbiosis. 1985;1:69–84. [Google Scholar]
- Von Uexkull HR, Mutert E. Global extent, development, and economic impact of acid soils. Plant Soil. 1995;171:1–15. [Google Scholar]
- Wasaki J, Omura M, Osaki M, Ito H, Matsui H, Shinano T, Tadano T. Structure of a cDNA for an acid phosphatase from phosphate-deficient lupin (Lupinus albus L.) roots. Soil Sci Plant Nutr. 1999;45:439–449. [Google Scholar]
- Watt M, Evans VR. Proteoid roots physiology and development. Plant Physiol. 1999;121:317–323. doi: 10.1104/pp.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]