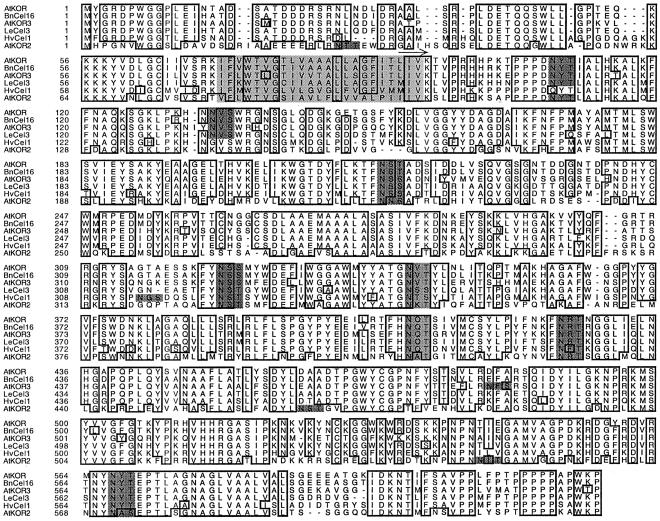
Figure 1.
Multiple alignment of deduced amino acid sequences of full-length plant membrane-anchored EGases. Abbreviations and Genbank accession nos. are as follows: Arabidopsis KOR (AtKOR, U37702), Arabidopsis KOR2 (AtKOR2, AC001229), Arabidopsis KOR3 (AtKOR3, AL078637), B. napus Cel16 (BnCel16, AJ242807), tomato Cel3 (LeCel3, U78526), and barley Cel1 (HvCel1, AB040769). Dots denote gaps to maximize alignment. Boxed residues are identical in at least five sequences. Dark-gray residues denote putative N-glycosylation sites among which six are conserved in the membrane-anchored EGase amino acid sequences. The core of the transmembrane domain is shown in light gray, and the catalytic domain of Cel16 expressed in P. pastoris is marked with an arrow above the sequence.