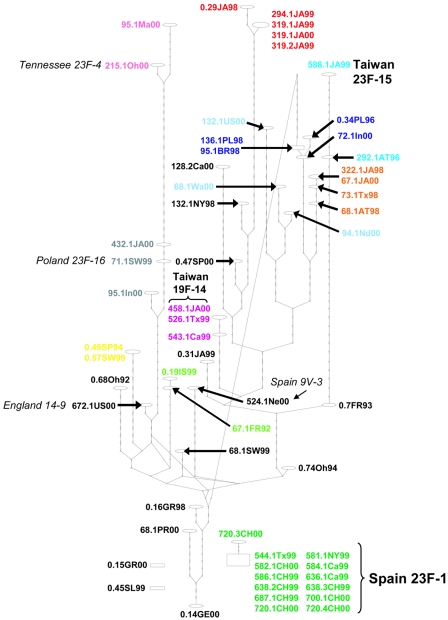
FIG. 1.
Statistical parsimony network (TCS) of the concatenated 8,163-bp, nonrecombinant housekeeping gene sequence alignment. Ovals represent different haplotypes, with the laboratory identifications for the isolates comprising that haplotype indicated adjacently. The number of steps (synonymous with intermediate or unsampled haplotypes) separating different haplotypes is represented by the nodes appearing on the branches. The rectangle represents the TCS determination of the haplotype most likely to be ancestral for this set of sequences. Spain 23F-1 is disconnected from the rest of the network because it requires too many steps (>19) to connect this group with the main body of the network. Clone names have been added, whenever this is possible, through a comparison to the allelic profiles listed for the 26 international clones on the PMEN website (http://www.sph.emory.edu/PMEN/index.html). Isolates that have alleles for the six MLST genes that we sequenced (ddl unknown) identical to a PMEN profile are indicated in boldface; those matching an international clone for five alleles (ddl unknown) are indicated in italics. Isolates of the same putative clone, based on our evolutionary criteria, are similarly colored in order to track the evolution of each of their fluoroquinolone target genes in Fig. 3. Our colored clonal groups, based on a consensus of closely adjacent haplotypes apparent in this housekeeping network and strongly supported monophyletic groups with short (or no) internodes in the case of phylogenies (Fig. 2), correspond very closely with the clonal group designation based on MLST criteria (29).