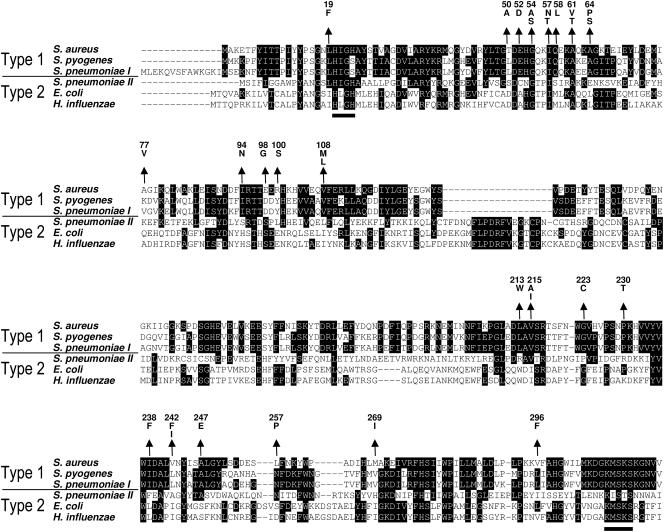
FIG. 4.
Alignment of MRS type 1 and type 2 amino acid sequences. Conserved residues are indicated by white letters on black background, and the H(M/I)GH and KMSKS motifs of the ATP-binding Rossman fold that are typically found in all class I tRNA synthetases are underlined. Arrows indicate the positions and point to amino acid changes in S. aureus mutants with decreased susceptibility to REP8839. Gaps introduced to maximize alignment are indicated by dashes.