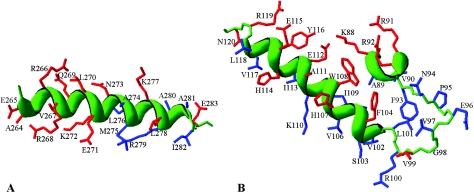
FIG. 5.
Structural models for σ32 residues 264 to 283 of region 4.2 (A) and for residues 88 to 120 of regions 2.2 to 2.4 (B). These models were rendered using DeepView-Swiss PDB Viewer version 3.7 by superimposing the sequences of E. coli σ32 onto the experimentally determined structures of T. aquaticus σA (3), as indicated on the figures. The justification for this simplified approach is derived from the observation that structure is remarkably well conserved among the σ70-like sigma factors. The side chains for which substitutions were introduced are colored red, and other residues are blue. The protein backbone is green, and for α-helices it is shown as a green ribbon.