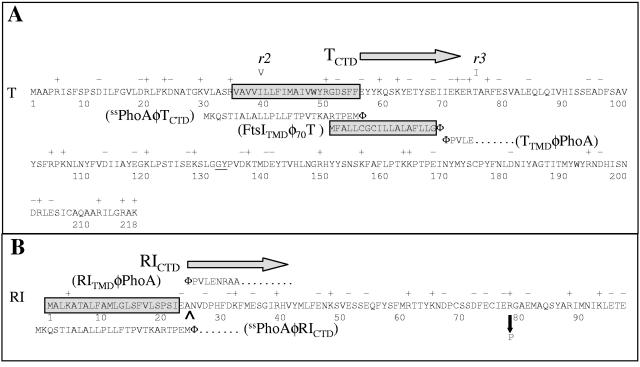
FIG. 2.
(A) Amino acid sequence of T and its derivatives. The predicted TMD of T is boxed and shaded. The rV mutants r2(I39V) and r3(T75I) are indicated above the sequence. In This, a hexahistidine tag (HHHHHHGG) was inserted between the residues G132 and Y133 (underlined). For the ssPhoAΦTCTD construct, the signal sequence of PhoA, shown below the T sequence, was substituted for residues 1 to 55 of T. For the FtsITMDΦT construct, the transmembrane domain of FtsI, shown in a shaded box below the T sequence, was substituted for residues 2 to 69 of T. For the TTMDφPhoA construct, residues 1 to 70 of T were substituted for residues 1 to 26 in the full-length primary gene product of phoA; the PhoA sequence begins with PVLE (from PhoA residue 27 on, as indicated). (B) Amino acid sequence of RI and its derivatives. The predicted signal sequence of RI is boxed, with the leader peptidase I cleavage site predicted by the SignalP program (http://www.cbs.dtu.dk/services/SignalP/) (2) indicated by a carat. In the c-myc-tagged version of RI, the c-myc tag (QKLISEEDL) was inserted after the terminal glutamate at position 97, while in the GFPΦRIcmyc constructs, the entire GFP sequence was inserted after the initial Met residue. For the ssPhoAφRICTD construct, the signal sequence of PhoA, shown below the RI sequence, was substituted for residues 1 to 24 of RI. For the RINTDφPhoA construct, the predicted signal sequence of RI replaced residues 1 to 26 in the uncleaved precursor to mature PhoA. The sole known rI missense mutation, R78P, is shown by a down arrow.