Abstract
The release of the 1000th complete microbial genome will occur in the next two to three years. In anticipation of this milestone, the Fellowship for Interpretation of Genomes (FIG) launched the Project to Annotate 1000 Genomes. The project is built around the principle that the key to improved accuracy in high-throughput annotation technology is to have experts annotate single subsystems over the complete collection of genomes, rather than having an annotation expert attempt to annotate all of the genes in a single genome. Using the subsystems approach, all of the genes implementing the subsystem are analyzed by an expert in that subsystem. An annotation environment was created where populated subsystems are curated and projected to new genomes. A portable notion of a populated subsystem was defined, and tools developed for exchanging and curating these objects. Tools were also developed to resolve conflicts between populated subsystems. The SEED is the first annotation environment that supports this model of annotation. Here, we describe the subsystem approach, and offer the first release of our growing library of populated subsystems. The initial release of data includes 180 177 distinct proteins with 2133 distinct functional roles. This data comes from 173 subsystems and 383 different organisms.
INTRODUCTION
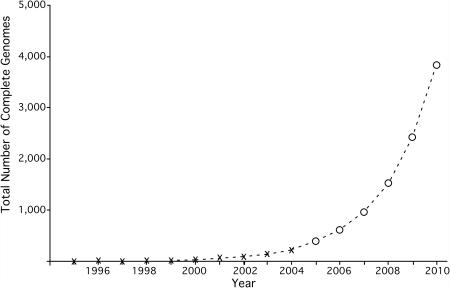
In the 10 years since the first complete bacterial genome was released in 1995 (1) there has been an exponential growth in the number of complete genomes sequenced. More than 200 complete genomes have been released, and based on past growth we anticipate that the 1000th genome will be sequenced at some point during 2007 (Figure 1). This rapid release of data reinforces the need for high-throughput annotation systems that provide reliable and accurate results.
Figure 1.
Accumulation of complete archaeal and bacterial genome sequences at NCBI 1994–2004, and prediction of the release of genomes through 2010. Data from http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi was extracted and plotted by year as shown with the crosses. Data from 2004–2010 is projected by the power law and is represented by open circles. At the current rate of growth, the 1000th complete microbial genome will be released in late 2007 or early 2008.
In response to these challenges the Fellowship for Interpretation of Genomes (FIG) launched the ‘Project to Annotate a 1000 Genomes’. The Project embodies a specific strategic view of how to approach high-throughput annotation: the effort is organized around subsytem experts, individuals who master the details of a specific subsystem and then analyze and annotate the genes that make up that given subsystem over an entire collection of genomes.
We argue that a subsystems based approach provides many benefits compared to more traditional techniques of genome annotation:
The analysis of a single subsystem over a large collection of genomes produces far more accurate annotations than the common approach of annotating the genes within a single organism. In fact the usual ‘gene-by-gene’ approach ensures that in most cases the individual annotating an entire genome lacks specific expertise related to the role of each gene.
The annotation of protein families rather than an organism at a time brings to bear specialized expertise and consequently leads to improvements over ‘gene-by-gene’ annotations of one genome. Just as the analysis of families offers a significant improvement over the annotation of individual genes, the analysis of sets of related protein families (i.e. those containing genes that make up a single biological subsystem) is more productive than the analysis of single families in isolation. Indeed, the fact that ‘The presence or absence of metabolic pathways and structures provides a context that makes protein annotation far more reliable’ (2) has now become clearly established.
It is both more straightforward and less error prone to automatically project annotations from a set of populated subsystems covering a diverse set of organisms than to project individual annotations using the existing automated pipelines. This is leading to the development of rule-based extension systems that will quite probably achieve superior accuracy (http://www.ebi.ac.uk/swissprot/Publications/dagstuhl.html).
A collection of annotations organized around specific subsystems covering a large number of diverse organisms represents a central resource for other bioinformatics efforts such as metabolic reconstruction, stoichiometric modeling and gene discovery (3).
This paper describes the subsystem-based approach to high-throughput genome annotation. The broad concepts of this approach are described and several examples of annotated subsystems are provided. Supplementary online material consisting of 173 subsystems has been released. Additionally, our open-source software for their creation and curation is provided.
WHAT IS A SUBSYSTEM
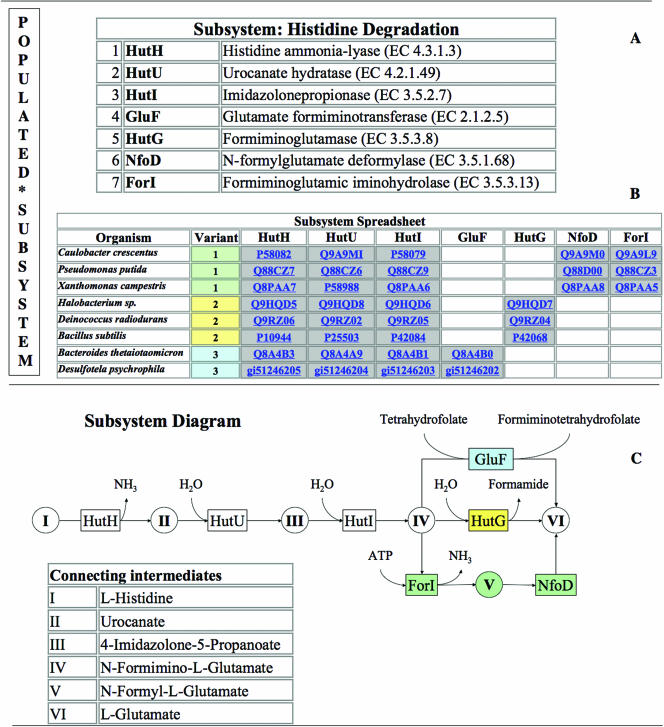
A subsystem is a set of functional roles that together implement a specific biological process or structural complex (Table 1). A subsystem may be thought of as generalization of the term pathway. Thus, just as glycolysis is composed of a set of functional roles (glucokinase, glucose-6-phosphate isomerase and phosphofuctokinase, etc.) a complex like the ribosome or a transport system can be viewed as a collection of functional roles. In practice, we put no restriction on how curators select the set of functional roles they wish to group into a subsystem, and we find subsystems being created to represent the set of functional roles that make up pathogenicity islands, prophages, transport cassettes and complexes (although many of the existing subsystems do correspond to metabolic pathways). The concept of populated subsystem is an extension of the basic notion of subsystem—it amounts to a subsystem along with a spreadsheet depicting the exact genes that implement the functional roles of the subsystem in specific genomes. The populated subsystem specifies which organisms include operational variants of the subsystem and which genes in those organisms implement the functional roles that make up the subsystem. Each column in the spreadsheet corresponds to a functional role from the subsystem, each row represents a genome, and each cell identifies the genes within the genome that encode proteins which implement the specific functional role within the designated genome (Figure 2).
Table 1.
Glossary
| Annotation | An unstructured text string associated with specific genes and/or proteins. |
| Clearinghouse | A site for publish-request type peer-to-peer exchange of subsystems in a system independent manner. |
| Functional role | An abstract function that a protein performs. Subsystems developers specify a single, precise text string to represent each functional role. |
| Functional variants | Different combinations of functional roles that represent distinct operational forms of a subsystem. |
| Missing gene | A gene, that is predicted to be present in the genome of an organism but has not been identified yet. |
| Populated subsystem | A subsystem along with a spreadsheet in which each column represents a functional role for the subsystem, each row represents a specific genome, and each cell contains those genes from the specific organism that have a subsystem connection to the specific functional role. |
| Product name | A short text string used to represent the function of the protein encoded by a gene. No constraints are placed on the strings used as product names, and it is common to see the same abstract function denoted by numerous similar expressions. |
| Protein family | A collection of proteins that were grouped by a curator. Proteins may be grouped based on domain structure, similarity, or some other characteristic. Proteins within a family may implement the same or multiple functional roles. |
| Subsystem | A Subsystem is a collection of functional roles, which together implement a specific biological process or structural complex. There is no distinction between metabolic subsystems and non-metabolic subsystems. |
| Subsystem connections | The set of functional roles that tie protein-encoding genes to different subsystems. Most protein encoding genes currently have a single subsystem connection. |
| Variant code | Numeric codes used to distinguish different functional variants. |
Figure 2.
Subsystem and Populated Subsytem. The Histidine Degradation Subsystem was used as an example to demonstrate relevant terms. (A) The subsystem comprises of 7 functional roles (e.g. Histidine ammonia-lyase (EC 4.3.1.3), Urocanate hydratase (EC 4.2.1.49) etc.). Together with the spreadsheet it becomes the ‘Populated subsystem’. (B) The Subsystem Spreadsheet is populated with genes from 8 organisms (simplified from the original subsystem) where each row represents one organism and each column one of the functional roles of the subsystem. Genes performing the specific functional role in the respective organism populate the respective cell. Gray shading of cells indicates proximity of the respective genes on the chromosomes. (C) The Subsystem Diagram illustrates the populated subsystem: key intermediates (circles with roman numerals), connected by enzymes (boxes with abbreviations matching the spreadsheet abbreviations) and reactions (arrows). There are three distinct variants of Histidine Degradation presented in this populated subsystem. Variant 1 (green shading) is present in Caulobacter crescentus, Pseudomonas putida and Xanthomonas campestris. N-Formimino-l-Glutamate (IV) is converted to l-Glutamate (VI) via N-Formyl-l-Glutamate (V) by enzymatic activities of Formiminoglutamic iminohydrolase (EC 3.5.3.13) (ForI) and of N-formylglutamate deformylase (EC 3.5.1.68) (NfoD). Variant 2 (yellow shading) is present in Halobacterium sp., Deinococcus radiodurans and Bacillus subtilis. In this variant the conversion from intermediate IV to VI is performed by Formiminoglutamase (EC 3.5.3.8) (HutG). Variant 3 (blue shading) is present in Bacteroides thetaiotaomicron and Desulfotela psychrophila. Here the Glutamate formiminotransferase (EC 2.1.2.5) (GluF) performs the conversion from intermediate IV to VI.
The act of populating the subsystem amounts to adding rows (i.e. genomes) to the spreadsheet.
Since these concepts are fundamental to our discussion we are illustrating them in Figure 2.
Note that each row in the spreadsheet has an associated variant code. The set of roles that make up the example subsystem include all of the functional roles needed to encode three common variants of the pathway. The variant codes distinguish three alternative means of converting N-formimino-l-glutamate to l-glutamate.
We have adhered to the position that experts encoding subsystems must decide exactly which functional roles to include (and exactly how to express each functional role), as well as what variant codes to use. We have restricted the use of two variant codes: 0 to represent work in progress and −1 to represent no operational variant.
A FRAMEWORK FOR DEVELOPING A PRECISE VOCABULARY FOR FUNCTIONAL ROLES
Controlled vocabularies have often been proposed in computer-assisted annotations and data mining (4,5). Subsystems technology supports the definition of a controlled vocabulary for gene function. Domain experts, by defining the functional roles that make up the subsystems that they curate, impose a precise vocabulary for assignment of function to the genes that implement the subsystem. Since the term ‘gene function’ has come to have several meanings, it is important to distinguish between four concepts:
A functional role is an abstract function such as ‘Aspartokinase (EC 2.7.2.4)’. Subsystems are sets of such abstract functions.
The notion of product name refers to a short text string that someone has used to represent the function of the protein encoded by a gene. There are no constraints on the strings used as product names, and it is common to see the same abstract function denoted by numerous similar expressions such as ‘Aspartokinase, Aspartokinase II, aspartate kinase’ etc.
By the term protein family we mean a collection of proteins that have been grouped by some curation team. The UniProt effort is producing one particularly valuable collection of families. Within that effort, the protein family represents a set of proteins that share a common domain structure. That is, they may actually implement the same or multiple functional roles. Within our work, there is no explicit concept of protein family; the closest notion would be ‘the set of genes within a single column of the spreadsheet in a populated subsystem’. However, a single column often contains proteins with distinct domain structure (e.g. both unifunctional and multifunctional proteins often occur within a single column), and in some cases genes encoding non-homologous proteins, which implement a single function have been included within a single column. We have developed tools to support comparison between protein families from a variety of sources and the proteins encoded by the genes in a single column in a populated subsystem. These comparisons are valuable but it is important to realize that we are producing sets of genes that encode proteins capable of implementing a single functional role, while the underlying restrictions on what make up a protein family often differ markedly from this notion.
The notation of annotation is often used to refer to an unstructured text string associated with specific genes and/or proteins.
To illustrate our use of these terms, consider the product name ‘Lysine-sensitive aspartokinase III’. It implements the functional role ‘Aspartokinase (EC 2.7.2.4)’, which a curator has included in the subsystem ‘Lysine_Biosynthesis_DAP_Pathway’. The curator may have well attached the annotation ‘Cassan et al., 1986 Nucleotide sequence of lysC gene encoding the lysine-sensitive aspartokinase III of Escherichia coli K12. Evolutionary pathway leading to three isofunctional enzymes, J. Biol. Chem., 261, 1052–1057’ for the respective E.coli K12 gene, justifying the use of this specific product name.
To this mix of concepts we add the notion subsytem connection. A gene can be connected to one or more functional roles, which induces connections to specific subsystems (those that contain the specific functional roles). In the example above it would be the connection to the subsystem ‘Lysine_Biosynthesis_DAP_Pathway’.
Although product names often include special properties (e.g. ‘thermostable’ or ‘lysine-sensitive’), and occasionally clues of function (e.g. ‘similar to death associated protein kinase’), subsystem connections unambiguously reference specific functional roles included in the definition of a subsystem.
Initially, the number of populated subsystems grew rapidly including numerous metabolic pathways, as well as non-metabolic subsystems ranging from flagella (http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=Flagellum&request=show_ssa, pathogenicity islands, http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=Mannose-sensitive_hemagglutinin_type_4_pilus&request=show_ssa), and secretory systems [http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=General_secretory_pathway_(Sec-SRP)_complex_(TC_3.A.5.1.1)&request=show_ssa] through complexes like the ribosome and proteosome. As both subsystems and the consequent subsystem connections matured there was considerable overlap between subsystems. Users developing subsystems on their own machines and sharing them through the clearinghouse exacerbated the differences in style, and hence conflicts between subsystems. For example, functional roles corresponding to the notion of aconitase exist in at least three distinct subsystems: the TCA cycle (http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=TCA_Cycle&request=show_ssa), the methylcitate cycle (http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=Methylcitrate_cycle&request=show_ssa), and glyoxylate synthesis (http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=Glyoxylate_Synthesis&request=show_ssa) developed independently by different curators. In at least one instance a curator wished to carefully distinguish three distinct forms of the enzyme. Initially each curator annotated the same protein-encoding genes with different functional roles, however this quickly became untenable—i.e. conflicts arose. To support uniform terminology required that the conflicts be detected, and be resolved by renaming functional roles to a consistent vocabulary employed consistently by all three subsystems. Rather than impose a centralized mechanism for resolving such conflicts, a completely decentralized approach was used.
To facilitate coordination and communication between end users, to aid with conflict resolution, and to eliminate redundancy, a multi-author website was developed using Wiki technology (http://www-unix.mcs.anl.gov/SEEDWiki/moin.cgi/MoinMoin). The subsystem bulletin board (http://www.theseed.org/wiki/moin.cgi/SubsystemBulletinBoard) provides an overview of the subsystems and highlights individual researcher's efforts. For a more detailed discussion of each of the subsystems, a Forum was developed using vBulletin technology (http://www.vbulletin.com/). The Forum (http://www.subsys.info) has subsystems separated by class, and each subsystem has a discussion arena for the deposition of comments, questions, suggestions and ideas. In addition to these resources, interactive conflict detection and resolution software was developed for the installation of subsystems in the SEED database.
Ultimately the success of our approach has been based on the good will and common desire to produce a consistent, precise vocabulary for functional roles, and we feel that this has worked well. It has produced a situation in which, at any given time, conflicts may exist because new subsystems are being developed or existing ones extended. But the attention of curators is being alerted to those instances by the development of tools that point to the conflicts. No centralized authority is being employed (although, in fact, on occasion curators do settle disagreements by consulting with outside experts). Conflicts can be of various types ranging from simple differences in spelling of functional roles to disagreements relating to specificity and numerous other issues. In all cases curators have reached settlements through discussions that lead to either consensus names or extended names. Once agreement has been reached and consistency established, changing the precise string of text that describes a functional role at some later point in time is trivial.
The result has been a vocabulary for functional roles that is precise, reasonably consistent, and rapidly improving. Our strategy for coupling this vocabulary with widely practiced ontologies such as GO will be to attach GO terms to each of the functional roles (inducing connections to genes via subsystem connections).
SUBSYSTEMS: A TECHNOLOGY INDEPENDENT OF ANNOTATION SYSTEMS
The subsystems technology described herein was developed with two primary goals in mind.
The first goal was to define a simple, portable text representation of a populated subsystem. This allowed populated subsystems to be exchanged, archived and updated over the Internet.
And the second goal to develop a clearinghouse where curators can publish populated subsystems for exchange with other users. The clearinghouse is available for direct querying from within a program (http://clearinghouse.theseed.org/) or via a web-browser (http://clearinghouse.theseed.org/clearinghouse_browser.cgi).
The development of this technology ensured that the subsystems information could be shared in a platform-independent manner, without requiring any centralized resource (such as a pathway collection). Any annotation environment can be developed or modified to support the creation and curation of subsystems using the clearinghouse (or, a local clearinghouse, if desired) as a repository.
THE SEED TECHNOLOGY TO SUPPORT SUBSYSTEMS
The SEED annotation environment is the first annotation environment that supports the creation, curation, population and exchange of subsystems. It supports publishing subsystems to a clearinghouse, and the downloading and installation of subsystems developed at other sites.
The SEED was developed by an international collaboration led by members of FIG and Argonne National Laboratory (6). The software is being made available as open source software released under the GNU public license (GPL) from the ftp site ftp://ftp.theseed.org/SEED.
Only a few enhancements would have to be added to any existing annotation system to support analysis of subsystems, and this functionality would extend existing software. The software would have to be extended to encode populated subsystems as objects and decode the populated subsystems as they are retrieved from the clearinghouse. Software would need to be included to publish and request populated subsystems from the clearinghouse. The software would have to be able to define the functional roles in initial subsystems, and to establish the subsystem connections between protein-encoding genes, functional roles and subsystems.
EXAMPLE POPULATED SUBSYSTEMS
Our populated subsystems were assembled into a single collection with a consistent formulation of functional roles and released via the web (http://www.theseed.org/Release1_Subsystems/index.html). An open source collection of software tools has been released via FTP ftp://ftp.theseed.org/SEED. To illustrate the advantages of subsystem based annotations over ‘traditional’ annotation systems several subsystems are described below:
Leucine Degradation and HMG-CoA Metabolism (http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=Leucine_Degradation_and_HMG-CoA_Metabolism&request=show_ssa)
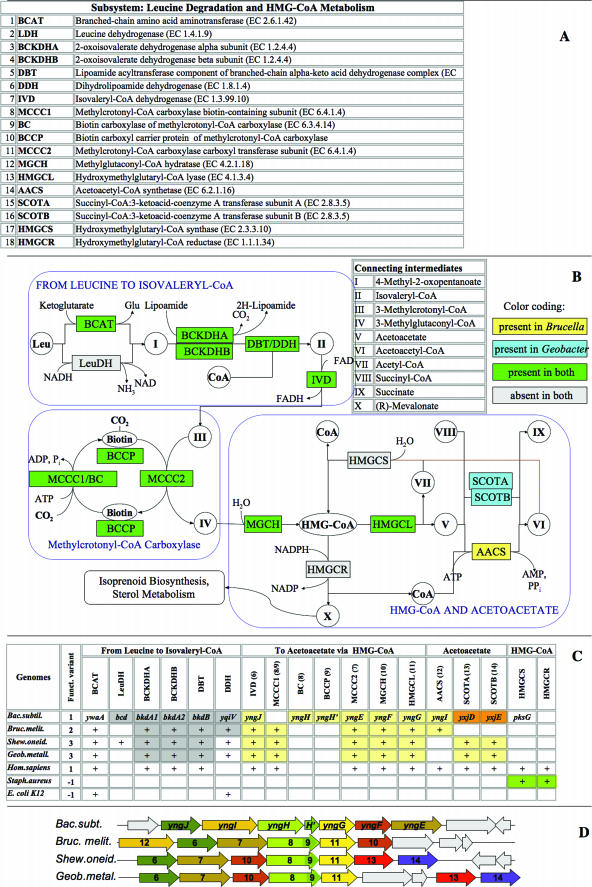
The populated subsystem presenting the leucine catabolism/HMG-CoA synthesis is depicted in Figure 3. An earlier analysis of some parts of this subsystem was presented elsewhere (7).
Figure 3.
Leucine Degradation and HMG-CoA Metabolism Subsystem. Functional roles, abbreviations, key intermediates and reactions in the pathway diagram are presented using the same conventions as in Figure 2. (A) Functional roles in the subsystem. (B) The Subsystem diagram shows the presence of genes assigned with respective functions for B.melitensis and G.metallireducens, using color-coded highlighting as explained in the panel. (C) Subsystem spreadsheet showing presence of genes with functions is shown by gene names for B.subtilis or by ‘+’ for all other genomes (modified from a regular SEED display showing all gene IDs). Highlighting by a matching color indicates proximity on the chromosome. (D) Clustering on the chromosome of genes involved in the Subsystem (large yellow cluster) demonstrated by alignment of the chromosomal contigs of respective genomes around a signature pathway gene, yngG. Homologous genes are shown by arrows with matching colors and numbers corresponding to functional roles in panel A. B.subtilis genes are marked by gene names. Other genes (not conserved within the cluster) are colored gray.
In humans leucine catabolism is coupled to sterol biosynthesis via a hydroxymethylglutaryl-coenzyme A (HMG-CoA) intermediate. The pathway is well characterized because defects in individual steps cause hereditary metabolic disorders like isovaleric acidemia, methylcrotonylglycinuria, methylglutaconic aciduria and 3-hydroxy-3-methylglutaric aciduria (8,9,10). Moreover, the human enzyme HMG-CoA reductase is a target in cardiovascular disease therapy because of its rate-limiting role in sterol biosynthesis (11). In contrast, only the early catabolic steps had been characterized in bacterial genomes—no genes were directly connected to enzymatic steps beyond isovaleryl-CoA (metabolite II in Figure 3B). Attempts to project from known eukaryotic genes based exclusively on homology searches produced ambiguous results because most of the enzymes in this pathway are members of large families of paralogs.
A combination of functional and genome context analysis, as depicted in the populated subsystem spreadsheet (Figure 3C) provided convincing evidence for the presence of the entire pathway of leucine catabolism in a number of diverse bacteria. A large conserved gene cluster containing reliable bacterial orthologs of two known human genes committed to this pathway was observed (Figure 3D). The gene yngH present in Bacillus and other bacteria is an ortholog of the human Methylcrotonyl-CoA carboxylase carboxyl transferase subunit (EC 6.4.1.4) while the neighboring gene yngG is an ortholog of HMG-CoA lyase (EC 4.1.3.4). This observation enabled the refinement of functional annotations for two additional bacterial genes in the same cluster (yngJ, an ortholog of Isovaleryl-CoA dehydrogenase (EC 1.3.99.10) and yngF, an ortholog of Methylcrotonyl-CoA carboxylase biotin-containing subunit (EC 6.4.1.4). Because these were weak homologs they could not be accurately characterized without considering the chromosomal neighborhood. The prediction (neither the bacterial nor the eukaryotic versions of methylglutaconyl-CoA hydratase were sequenced at that point) of yngG performing this function was projected from Bacillus to the human homolog. Later this prediction was proven correct by two independent publications that provided the experimental verification of the function encoded by this human gene (12,13).
Another functional inference from the analysis of this subsystem was a connection between leucine catabolism and acetoacetate metabolism (as illustrated in Figure 3B). This observation suggested a physiologically relevant extension of the HMG-CoA subsystem beyond its traditional boundaries. Two forms of yngF (encoding the methylcrotonyl-CoA carboxylase biotin-containing subunit (EC 6.4.1.4) were observed—the most common form, a fusion of biotin carboxylase and a C-terminal biotin carboxylase carrier protein domain and a rare form, in which the biotin carboxylase and the downstream biotin carboxylase carrier protein-encoding gene are separate (as in B.subtilis). The subsystems approach allows for different variants of enzymes as shown in Figure 3.
Panels B and C in Figure 3 illustrates the analysis of functional variants of a subsystem. Most of the subsystem protein-encoding genes are conserved in those species that have a functional (‘nonzero’) variant. However, E.coli and Staphylococcus aureus do not have a functional variant leading to the inference that they are incapable of catabolizing leucine using this pathway. Consequently, they were marked ‘−1’ in the subsystem spreadsheet (Figure 3C). A distinction between the functional variants 1–3 was made based on the downstream component of the subsystem: the alternative routes of conversion of acetoactetate to succinate (intermediate V in Figure 3B). This was either via Succinyl-CoA:3-ketoacid-coenzyme A transferase subunits A and B (EC 2.8.3.5) (variant 2; e.g. Brucella melitensis) or via Acetoacetyl-CoA synthetase (EC 6.2.1.16) (variant 3; e.g. Geobacter metallireducens and Shewanella oneidensis). Both routes were possible in variant 1, as exemplified by both human and B.subtilis, although clustering on the chromosome suggests that in the latter species an AACS-dependent reaction may be preferred or co-regulated with the other components of the subsystem.
This example illustrates how prokaryotic chromosomal clustering can influence the interpretation of pathways, prediction of missing genes and projection of annotations between prokaryotic and eukaryotic genes. The observations also contributed to interpretation of the evolutionary history of a large and diversified group of proteins. More such examples have been published elsewhere (3,14).
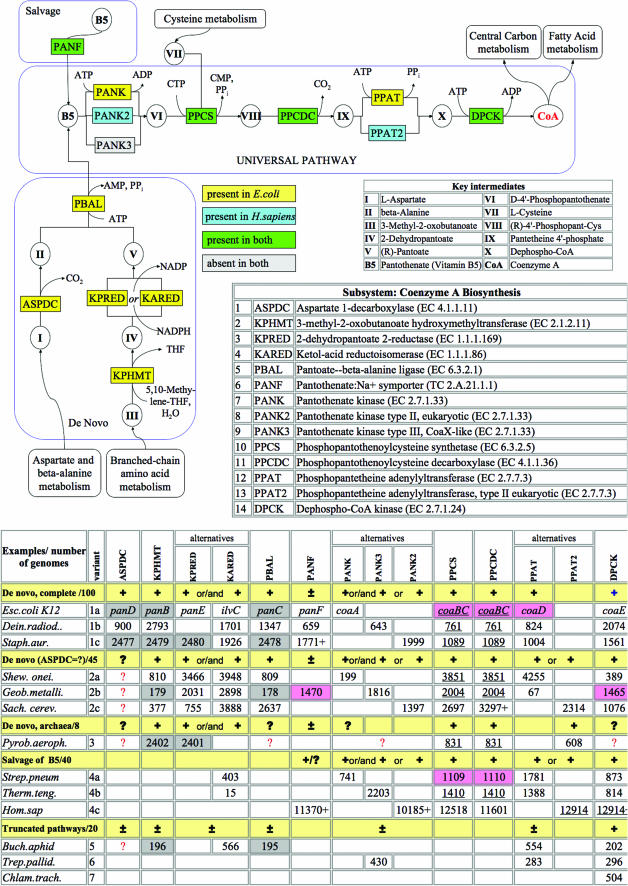
Coenzyme A biosynthesis subsystem (http://www.theseed.org/annocopy/FIG/subsys.cgi?ssa_name=Coenzyme_A_Biosynthesis&request=show_ssa)
Coenzyme A (CoA) is a universal and essential cofactor in all forms of cellular life (15). Earlier bioinformatics analysis of CoA biosynthesis revealed a number of interesting variations between species (3,16,17). In the respective SEED subsystem (see Figure 4), this analysis was extended to >250 diverse genomes. A five-step pathway from pantothenate (vitamin B5) to CoA is the universal component of the subsystem conserved in the majority of species. The most variable aspect of this pathway is pantothenate kinase (PANK). Three non-orthologous forms of PANK are presently known, and, in some cases, two alternative forms are present in the same organism. A recently identified and characterized CoaX-like (type III) pantothenate kinase (PANK3) appears to be more common in the bacterial world than the ‘classic’ PANK1 (18). Nevertheless, in most genomes, homologs of PANK3 have misleading annotations (e.g. ‘BVG accessory factor’). The populated subsystem allows one to suggest reliable annotations for these proteins in many bacterial genomes, strongly supported by the strict requirement of PANK for CoA biosynthesis. The eukaryotic-like PANK2 was predicted (19) and subsequently verified (20) as the only PANK in all Staphylococcus species.
Figure 4.
CoA Biosynthesis Subsystem. Functional roles, abbreviations, key intermediates and reactions in the pathway diagram are presented using the same conventions as in Figure 2. Background colors in the diagram illustrate the comparison of subsystem variants by highlighting functional roles asserted in two organisms: E.coli (yellow) and H.sapiens (blue). Shared functional roles are highlighted green. The lower panel is a modification of the subsystem spreadsheet. It shows a classification of major subsystem variants representing a substantially different reaction topology revealed by semi-automated graph analysis as described in (21). Selected genomes unambiguously associated with each variant are shown after variant description (e.g. De novo, complete/100). Patterns of functional roles which constitute each functional variant are generalized by: ‘+’, presence of a gene (for a given role) is required; ‘±’, optional; ‘?’, function is inferred by pathway analysis but a gene is unknown or ‘missing’ (i.e. can not be located by similarity). Typical sub-variants characterized by the same topology but relying on alternative (non-orthologous) forms of specific enzymes (e.g. PANK) are illustrated by the following genomes: E.coli K12 [NCBI taxonomy ID 83333.1], D.radiodurans R1 [243230.1], S.aureus subsp. aureus N315 [158879.1], S.oneidensis MR-1 [211586.1], G.metallireducens [28232.1], Saccharomyces cerevisiae [4932.1], P.aerophilum str. IM2 [178306.1], Streptococcus pneumoniae R6 [171101.1], Thermoanaerobacter tengcongensis [119072.1], H.sapiens [9606.2], B.aphidicola str. APS (Acyrthosiphon pisum) [107806.1], Treponema pallidum subsp. pallidum str. Nichols [243276.1] and Chlamydia trachomatis D/UW-3/CX [272561.1]. Genes assigned with respective functional roles are shown by SEED unique IDs for all illustrated genomes (except E.coli where common gene names are used). Matching background colors highlight genes that occur close to each other on the chromosome.
A possible fourth non-orthologous form of PANK can be inferred from the analysis of Archaea. The candidate for the missing archaeal PANK is a member of the GHMP kinase family which clusteres on the chromosome with several other CoA biosynthetic genes in some Archaea (i.e. PAE3407 of Pyrobaculum aerophilum). Another conserved family (represented by PAE1629 of P.aerophilum) may fulfill the role of dephospho-CoA kinase (DPCK), which is still ‘missing’ in all Archaea. This conjecture is based on a long-range sequence similarity with bacterial and eukaryotic enzymes (as suggested by the tentative annotation of COG0237 at NCBI http://www.ncbi.nlm.nih.gov/COG/old/palox.cgi?COG0237).
Both functional predictions [also suggested by (17)] require experimental verification. Among other problems within this subsystem is a missing aspartate decarboxylase in a number of genomes with an otherwise complete set of genes for the de novo synthesis.
Several examples illustrating major functional variants of the subsystem are outlined in Figure 4. An algorithm of semi-automated variant classification and a brief analysis of the key operational variants of CoA biosynthesis were recently published (21). Most species implement either complete de novo biosynthesis (variants 1–3) or a five-step pantothenate salvage (variant 4). A relatively small group of bacteria, most notably obligate intracellular pathogens and symbionts, display a variety of truncated pathways. For example, a disrupted pattern (missing PANK, PPCS and PPCDC) observed in Buchnera aphidicola suggests a possible metabolic exchange between this endosymbiont and the aphid host cell. According to this hypothesis, pantothenate produced but not utilized by B.aphidicola may be fed directly into the universal pathway of the host. The latter may pay back by providing a phosphopantetheine intermediate required for the last two steps of CoA synthesis in B.aphidicola. Several other interesting aspects of this subsystem are discussed in the supplementary materials (http://www.theseed.org/Release1_Subsystems/index.html).
Ribosomal proteins (http://www.theseed.org/SubsystemStories/Ribosomal_proteins/abstract.htm)
Historically, ribosomal proteins were identified in several important experimental organisms, including E.coli, Bacillus species, yeast, rat and Halobacterium. In each case, a unique nomenclature was developed. More recently, several groups sought unified nomenclatures given the availability of so many sequences. In the cases of Bacteria and Eukarya, these efforts were hugely successful. The most problematic aspects of the conventions were (i) the failure to uniformly indicate whether a given label is based upon the bacterial or the eukaryal numbering, and (ii) the linking of equivalent eukaryal and bacterial terms. There are only two proteins (S3 and L3) for which the bacterial and eukaryal numbers are the same. This created a particularly confusing situation when the bacterial nomenclature was applied to Archaea, except when no bacterial homolog existed, in which case the eukaryal label was applied.
To address these problems a dual labeling was applied in which bacterial proteins were given the bacterial label (always explicitly including the ‘p’, e.g. S5p), followed by the designation of the corresponding eukaryal protein in parentheses (always with the explicit ‘e’, e.g. S2e). Similarly, in the case of eukarya, the eukaryal protein designation is given first, followed by the bacterial label in parentheses. In the case of Archaea, in all but a few cases the proteins are clearly of the eukaryal genre, and the eukaryal term is given first. One of the most important consequences of this nomenclature is that a text-based search is always unambiguous as to whether the bacterial or eukaryal numbering is desired. For example, a search for L11p will return bacterial L11 and eukaryal L12, but not bacterial L5 (the equivalent of eukaryal L11). A second key decision was to use the terms LSU and SSU to distinguish the subunits, rather than 30S, 40S, 50S and 60S. In addition to further unifying the nomenclature, it avoids two key sources of confusion. Several eukaryal ribosomes (especially organellar ribosomes) have been assigned to ‘non-standard’ sizes. Thus, searching for 50S and/or 60S was not sufficient to ensure that all ribosomes were distinguished. But more importantly, it avoids the temptation to use 50S to designate the LSU of a eukaryal mitochondrial ribosome. Instead, we have explicitly identified all organellar proteins by ‘mitochondrial’ or ‘chloroplast’.
The development of this nomenclature demonstrated the power of the subsystems approach for encoding non-metabolic pathways, and the utility of functional roles in describing a controlled vocabulary for gene product function.
THE IMPACT OF POPULATED SUBSYSTEMS
As demonstrated by the examples above, populated subsystems can be used to support two broad categories of research: advancing research in the populated subsystems themselves and addressing numerous fundamental problems within bioinformatics.
It is important to note that there are large and ongoing efforts that address similar objectives—most notably the KEGG (http://www.genome.jp/kegg/kegg2.html) (22,23), GO (http://www.geneontology.org/) (5) and MetaCyc (http://metacyc.org/) (24) projects. These represent substantial projects, and we have in many ways built upon their work. Perhaps, the most obvious difference between our work and these projects is that we have made it possible for all researchers to immediately develop detailed encodings of their particular area of expertise, to make these new encodings available to the research community, and to import the work of others in constructing a customized collection of subsystems covering their specific needs. This radically decentralized effort offers a different set of incentives for domain experts to participate, which is precisely what will be needed to improve existing annotations.
The primary utility of annotated subsystems relates to the fact that a populated subsystem often supports substantially more accurate assignments of function to genes.
In addition the analysis of the populated subsystem allows one to arrive at a precise notion of which forms (i.e. which variants) of the subsystem exist in which organisms.
Further, the spreadsheet included in an populated subsystem often makes it vividly clear that a gene implementing a specific functional role is very likely to exist, even though it has not yet been identified. These so-called missing gene problems occur with surprising frequency. In the two metabolic examples presented in this paper and in various instances published in the Supplemental Material we show in detail a few instances in which conjectures could easily be formulated once the actual presence of a missing gene had been identified.
Finally, the presence of an extensive set of annotated subsystems lays the foundation for an accurate characterization of the metabolic network present in each organism.
The existence of a collection of populated subsystems also has an impact on a number of important topics in bioinformatics:
Over and over as we performed our analysis we found that genes that appeared to actually be missing in an annotated subsystem were, in fact, present within an open reading frame (ORF), but eluded identification by a gene-calling algorithm. For the functional roles represented in populated subsystems, it becomes possible to directly search for instances of these roles in cases in which there is reason to believe that such a gene must exist.
Once ORFs containing genes have been identified, the problem of accurately identifying the start of the gene remains. The most successful attempts have been based on alignments. We argue that use of genes that are both similar and believed to implement the same functional role will lead to substantial improvements over existing estimates. A team at Middle Tennessee State University has brought up a website (http://torvalds.cs.mtsu.edu/cgi-bin/starts/starts.cgi) with initial results.
The search for regulatory sites in upstream regions of related genes has often led to success (25). Regulon analysis in combination with other techniques of comparative genomics was allowed to improve interpretation and to generate functional predictions in a number of metabolic subsystems (26,27). With the release of our initial set of annotated subsystems, we are making data available to support such analysis. For each annotated subsystem, we are providing sequences of upstream regions for each prokaryotic genome. Each sequence contains 300 bp of upstream sequence depicting the boundary of the adjacent gene (delimiting the intergenic gap), as well as 100 bp of the gene sequence itself.
The development of carefully curated protein families has historically been a key goal of bioinformatics for obvious reasons. The limitations of existing formulations relate to ambiguities in function assignment, a problem that is directly addressed by annotated subsystems. We have used this initial collection to create a list of refinements of UniProt annotations, and we will work to make sure that our analysis directly supports both the UniProt and other efforts to produce clean, comprehensive collections of protein families.
Some of the most successful applications of bioinformatics technology relate to context analysis (3,28,29). In numerous cases, the clues that led to conjectures of function were based on the fact that related genes tend to cluster on prokaryotic chromosomes, tend to fuse and to co-occur. The annotated subsystems offer a framework for establishing the statistical properties needed to effectively exploit these tendencies.
The long-term goal of the subsystems approach is to bring every subsystem to a point where it has been carefully curated by one or more experts in the biological process encoded by the given subsystem. This approach will lead to the construction of an accurate phylogenetic context for each of the proteins within the subsystem, resulting in the ability to accurately trace the evolutionary histories of the catalytic domains that make up each subsystem [for a detailed illustration of this style of analysis, see ref. (30)].
Subsystems have also provided an approach for understanding the metabolism of environmental samples. A comparison of statistically significantly different subsystems present in different large environmental (metagenome) samples yielded unprecedented insights into the biology of these environments and lead to the generation of novel hypotheses to be tested by field biologists (R. Edwards, unpublished data).
THE RELEASE
Concurrent with the publication of this paper, an initial snapshot release of our collection of populated subsystems (which was a subset of those available via the SEED clearinghouse) was made. This subset is available in a format that makes the data easily accessible for use in other systems or as raw data. The current release of 173 populated subsystems is available without restriction via the web. The supplementary online subsystems material includes three main components:
A set of 48 example subsystems. These constitute examples that have been curated in somewhat more detail and have led to interesting conjectures or research results in a number of cases. For each of these examples, we include the complete subsystem ‘frozen’ at the time of release, abstracts, presentations or summaries providing more detail about the subsystem and suitable for classroom use or lectures.
A set of 173 populated subsystems ‘frozen’ at the time of release that cover a large swath of central metabolism and other cellular processes.
Links to the current status of each subsystem. Each of the subsystems is continually being curated and populated as new genomes are added to the SEED and new comparisons become available. These links provide access to the most up-to-date annotations.
Each provided sequence was packaged with as many IDs as possible. For example, identifiers from FIG, UniProt, KEGG and NCBI (including GI number, gene number, UI or RefSeq ID), as well as identifiers from sequencing laboratories were included to ensure portability. The SEED release is itself open source software and can be acquired via FTP ftp://ftp.theseed.org/SEED. The system was developed to run on both Mac OSX systems and Linux systems.
CONCLUSIONS
Within 2–3 years we will all have access to over a thousand sequenced genomes. This data will grow to become the central resource in modern biology. Annotating this collection is the core challenge of modern bioinformatics. In this paper we describe a new approach to annotation based on idea of subsystems that promises to dramatically improve the quality and utility of annotations. This approach is central to the Project to Annotate 1000 genomes and has been implemented in a suite of tools for genome annotation. The approach and technology provide one way to involve many domain experts in the genome annotation process. The technology for developing these subsystems now exists, the technologies for supporting automated addition of new genomes to the collection of populated subsystems is now being developed, and the initial collection is being made available to the research community.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the Fellowship for Interpretation of Genomes.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M., et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Haft D.H., Selengut J.D., Brinkac L.M., Zafar N., White O. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics. 2005;21:293–306. doi: 10.1093/bioinformatics/bti015. [DOI] [PubMed] [Google Scholar]
- 3.Osterman A., Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 4.Overbeek R., Larsen N., Smith W., Maltsev N., Selkov E. Representation of function: the next step. Gene. 1997;191:GC1–GC9. doi: 10.1016/s0378-1119(96)00854-2. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overbeek R., Disz T., Stevens R. The SEED: a peer-to-peer environment for genome annotation. Commun. ACM. 2004;47:46–51. [Google Scholar]
- 7.Overbeek R., Devine D., Vonstein V. Curation is forever: comparative genomics approaches to functional annotation. Targets. 2003;2:138–146. [Google Scholar]
- 8.Tanaka K., Ikeda Y., Matsubara Y., Hyman D.B. Molecular basis of isovaleric acidemia and medium-chain acyl-CoA dehydrogenase deficiency. Enzyme. 1987;38:91–107. doi: 10.1159/000469195. [DOI] [PubMed] [Google Scholar]
- 9.Weyler W., Sweetman L., Maggio D.C., Nyhan W.L. Deficiency of propionyl-Co A carboxylase and methylcrotonyl-Co A carboxylase in a patient with methylcrotonylglycinuria. Clin. Chim. Acta. 1977;76:321–328. doi: 10.1016/0009-8981(77)90158-9. [DOI] [PubMed] [Google Scholar]
- 10.Gibson K.M., Lee C.F., Hoffmann G.F. Screening for defects of branched-chain amino acid metabolism. Eur. J. Pediatr. 1994;153:S62–67. doi: 10.1007/BF02138780. [DOI] [PubMed] [Google Scholar]
- 11.Marz W., Wieland H. HMG-CoA reductase inhibition: anti-inflammatory effects beyond lipid lowering? Herz. 2000;25:117–125. doi: 10.1007/pl00001949. [DOI] [PubMed] [Google Scholar]
- 12.Loupatty F.J., Ruiter J.P., L I.J, 1st, Duran M., Wanders R.J. Direct nonisotopic assay of 3-methylglutaconyl-CoA hydratase in cultured human skin fibroblasts to specifically identify patients with 3-methylglutaconic aciduria type I. Clin. Chem. 2004;50:1447–1450. doi: 10.1373/clinchem.2004.033142. [DOI] [PubMed] [Google Scholar]
- 13.Ly T.B., Peters V., Gibson K.M., Liesert M., Buckel W., Wilcken B., Carpenter K., Ensenauer R., Hoffmann G.F., Mack M., et al. Mutations in the AUH gene cause 3-methylglutaconic aciduria type I. Hum. Mutat. 2003;21:401–407. doi: 10.1002/humu.10202. [DOI] [PubMed] [Google Scholar]
- 14.Jordan I.K., Henze K., Fedorova N.D., Koonin E.V., Galperin M.Y. Phylogenomic analysis of the Giardia intestinalis transcarboxylase reveals multiple instances of domain fusion and fission in the evolution of biotin-dependent enzymes. J Mol. Microbiol. Biotechnol. 2003;5:172–189. doi: 10.1159/000070268. [DOI] [PubMed] [Google Scholar]
- 15.Begley T.P., Kinsland C., Strauss E. The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 2001;61:157–171. doi: 10.1016/s0083-6729(01)61005-7. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes S.Y., Scholle M.D., D'Souza M., Bernal A., Baev M.V., Farrell M., Kurnasov O.V., Daugherty M.D., Mseeh F., Polanuyer B.M., et al. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genschel U. Coenzyme A biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol. Biol. Evol. 2004;21:1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- 18.Brand L.A., Strauss E. Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J. Biol. Chem. 2005;280:20185–20188. doi: 10.1074/jbc.C500044200. [DOI] [PubMed] [Google Scholar]
- 19.Daugherty M., Polanuyer B., Farrell M., Scholle M., Lykidis A., de Crecy-Lagard V., Osterman A. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 2002;277:21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- 20.Choudhry A.E., Mandichak T.L., Broskey J.P., Egolf R.W., Kinsland C., Begley T.P., Seefeld M.A., Ku T.W., Brown J.R., Zalacain M., et al. Inhibitors of pantothenate kinase: novel antibiotics for staphylococcal infections. Antimicrob. Agents Chemother. 2003;47:2051–2055. doi: 10.1128/AAC.47.6.2051-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y., Osterman A., Overbeek R., Godzik A. Automatic detection of subsystem/pathway variants in genome analysis. Bioinformatics. 2005;21:478–486. doi: 10.1093/bioinformatics/bti1052. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieger C.J., Zhang P., Mueller L.A., Wang A., Paley S., Arnaud M., Pick J., Rhee S.Y., Karp P.D. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelfand M.S., Novichkov P.S., Novichkova E.S., Mironov A.A. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 2000;1:357–371. doi: 10.1093/bib/1.4.357. [DOI] [PubMed] [Google Scholar]
- 26.Rodionov D.A., Vitreschak A.G., Mironov A.A., Gelfand M.S. Comparative genomics of thiamin biosynthesis in procaryotes: new genes and regulatory mechanisms. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 27.Rodionov D.A., Mironov A.A., Gelfand M.S. Conservation of the biotin regulon and the BirA regulatory signal in eubacteria and archaea. Genome. Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin E.V., Galperin M.Y. Sequence-Evolution-Function: Computational Approaches in Comparative Genomics. Boston: 1st Edn Kluwer Academic Publishers; 2002. [PubMed] [Google Scholar]
- 29.Huynen M.A., Snel B., von Mering C., Bork P. Function prediction and protein networks. Curr. Opin. Cell. Biol. 2003;15:191–198. doi: 10.1016/s0955-0674(03)00009-7. [DOI] [PubMed] [Google Scholar]
- 30.Xie G., Keyhani N.O., Bonner C.A., Jensen R.A. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 2003;67:303–342. doi: 10.1128/MMBR.67.3.303-342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]