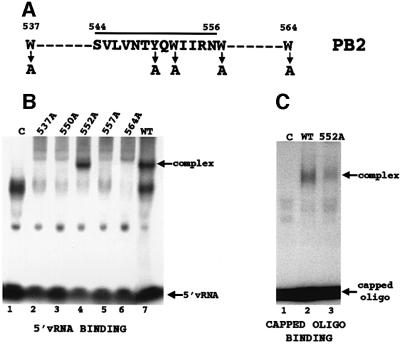
Fig. 2. Identification of the polymerase sequence that binds the 5′ terminal cap structures of RNAs. (A) Amino acid sequence of the peptide (amino acids 544–556) that cross-links to the capped, thio U-containing oligonucleotide containing 32P only in its 5′ cap structure. The positions of neighboring tryptophans (Ws) are also shown. The four tryptophans and the single tyrosine that were individually replaced with alanine are denoted by arrows. (B) Binding of 5′ vRNA (labeled at its 3′ end with 32pCp) to polymerase complexes containing a wild-type PB2 protein or one of the mutant PB2 proteins. The same amount (2.5 µg) of each nuclear extract was added. Polymerase–5′ vRNA complexes were separated from unbound 5′ vRNA by electrophoresis on 4% non-denaturing gels. Lane 1 (C), labeled 5′ vRNA incubated in the presence of a nuclear extract from cells infected with a vaccinia virus vector expressing T7 RNA polymerase. The virus-specific complex is denoted. The species that migrate faster than the virus-specific complex, which are also found in the absence of polymerase complexes, i.e. in lane 1 (C), vary in amount in different experiments. (C) Binding of a capped RNA fragment to polymerase complexes containing a wild-type PB2 protein or the mutant PB2 protein with an alanine substitution at position 552. The same amount (2.5 µg) of each nuclear extract was added. Subsequent to the binding of unlabeled 5′ vRNA to the polymerase complexes, the 13-nucleotide-long capped AlMV RNA 4 RNA fragment containing an m7G32pppGm 5′ end (10 ng, 1 × 105 c.p.m.) was incubated with the wild-type (lane 2) or the mutant polymerase complexes (lane 3). The capped RNA–polymerase complexes were separated from free capped RNA fragments by electrophoresis on 4% non-denaturing gels. The arrows denote the positions of free capped RNA fragments and of the capped RNA–polymerase complexes. The species that migrate faster than the virus-specific complex, which are also found in the absence of polymerase complexes (lane 1), vary in amount in different experiments. The amounts of the cap-binding complexes were quantitated using a phosphoimager. Lane 1 (C), capped RNA fragment incubated with a nuclear extract from cells infected with a vaccinia virus vector expressing T7 RNA polymerase.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.