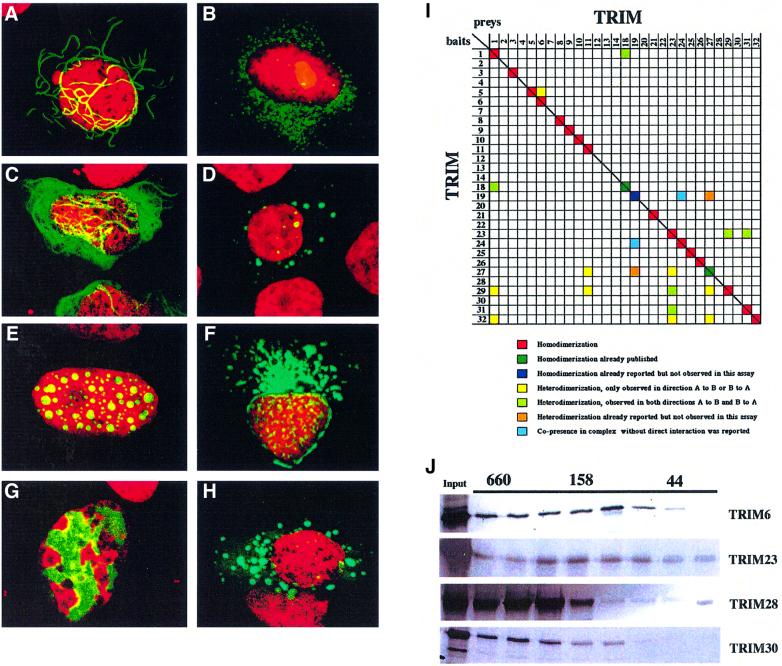
Fig. 4. TRIM proteins homomultimerize and associate with specific subcellular structures. Subcellular localization of TRIM29 (A), TRIM4 (B), TRIM2 (C), TRIM5 (D), TRIM8 (E), TRIM13 (F), TRIM28 (G) and TRIM9 (H). U2OS and HeLa cells were transfected with plasmids encoding EGFP–TRIM fusions. All TRIM proteins localized to particular subcellular compartments: TRIM29, to cytoplasmic ribbon-like structures; TRIM2, to cytoplasmic filaments; TRIM4, 5 and 9, to ‘cytoplasmic bodies’; TRIM8, to specific nuclear bodies; TRIM13, around the nucleus; and TRIM28, to specific chromatin regions. (I) Interaction-mating matrix of 32 TRIM ORFs fused to the LexA binding domain (bait) or to the B42-SV40 NLS-HA-tag domains (preys). Color coding is as follows: red squares, homodimerization; dark green squares, homodimerization previously reported and detected in the present assay; dark blue squares, homodimerization previously reported and not detected in the present assay; yellow squares, heterodimerization detected in either bait-X/prey-Y or bait-Y/prey-X orientation; light green squares, heterodimerization detected in both bait-X/prey-Y and bait-Y/prey-X orientations; orange squares, heterodimerization previously reported and not detected in either bait-X/prey-Y or bait-Y/prey-X orientation; light blue squares, co-presence without direct interaction in a protein complex reported (Grignani et al., 1996; Cao et al., 1997, 1998; Cainarca et al., 1999; Zhong et al., 1999a). (J) Superose 6 gel filtration of in vitro translated TRIM6, 23, 28 and 30 proteins. Their elution peaks correspond to formation of homomultimeric complexes. Elution of proteins of known molecular weight (kDa) is indicated above.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.