Abstract
In the endoplasmic reticulum (ER), disulfide bonds are simultaneously formed in nascent proteins and removed from incorrectly folded or assembled molecules. In this compartment, the redox state must be, therefore, precisely regulated. Here we show that both human Ero1-Lα and Ero1-Lβ (hEROs) facilitate disulfide bond formation in immunoglobulin subunits by selectively oxidizing PDI. Disulfide bond formation is controlled by hEROs, which stand at a crucial point of an electron-flow starting from nascent secretory proteins and passing through PDI. The redox state of ERp57, another ER-resident oxidoreductase, is not affected by over-expression of Ero1-Lα, suggesting that parallel and specific pathways control oxidative protein folding in the ER. Mutants in the Ero1-Lα CXXCXXC motif act as dominant negatives by limiting immunoglobulin oxidation. PDI-dependent oxidative folding in living cells can thus be manipulated by using hERO variants.
Keywords: endoplasmic reticulum/folding/immunoglobulins/oxidoreductases/redox
Introduction
To assist the folding of proteins destined for the secretory pathway and extracellular space, the endoplasmic reticulum (ER) of eukaryotic cells contains a vast array of chaperones and enzymes (Gething and Sambrook, 1992; Ellgaard et al., 1999). As many secretory proteins contain disulfide bonds, a central role is played by ER-resident oxidoreductases, such as PDI, ERp72 (CaBP1), ERp57 and P5 (CaBP2). Recently, PDI and ERp57 have been shown to react directly and selectively with different substrate proteins and, therefore, to participate in distinct oxidative pathways (Molinari and Helenius, 1999, 2000). Preferential import of oxidized glutathione (GSSG) from the cytosol has been proposed to generate oxidizing conditions in the ER (Hwang et al., 1992). However, the redox state must be precisely regulated within the ER, as in this organelle disulfide bonds are simultaneously formed in nascent proteins and removed from incorrectly folded or assembled polypeptides (Helenius et al., 1992). The process of disulfide bond formation has been elegantly reconstructed in Saccharomyces cerevisiae. PDI molecules that are reduced after donating a disulfide bond to cargo proteins are re-oxidized by Ero1p, the product of the essential ERO1 gene (Frand and Kaiser, 1999). Ero1p has been shown to oxidize PDI and another ER oxidoreductase, Mpd2p (Frand and Kaiser, 1999) and to require flavin adenine dinucleotide as a cofactor (Tu et al., 2000), but the mechanisms that control its re-oxidation in vivo remain to be elucidated. The phenotype of yeast cells lacking functional Ero1p can be alleviated by diamide (Pollard et al., 1998) and suppressed by disruption of the glutathione synthase gene (Cuozzo and Kaiser, 1999). These findings suggest that Ero1p serves mainly as an oxidant, and that GSH may act as a buffer of the oxidant power of Ero1p.
We cloned two human members of the ERO1 family, ERO1-Lα and ERO1-Lβ (hEROs), which show different tissue distribution and transcriptional regulation. Only ERO1-Lβ is induced during the unfolded protein response (Pagani et al., 2000). Nevertheless, both genes complement the ero1-1 yeast mutant (Cabibbo et al., 2000; Pagani et al., 2000). Ero1-Lα can be found in mixed disulfides with PDI in mammalian cells (Benham et al., 2000) confirming its involvement in the pathways of disulfide bond formation.
To investigate and compare the function of Ero1-Lα and Ero1-Lβ, we monitored the rate and efficiency of oxidative folding of two immunoglobulin subunits, J and κ chains, in the ER of mammalian cells over-expressing either Ero1-Lα or Ero1-Lβ. Our results show that both oxidireductins facilitate disulfide bond formation in cargo proteins. The two hEROs form mixed disulfides with PDI and other unidentified proteins. Covalent complexes with ERp72, ERp57 and P5 are not detected, suggesting that selective protein–protein interactions underlie the control of the redox state in the ER. Certain Ero1-Lα mutants act as dominant negatives; therefore, the activity of hEROs seems to play a pivotal role in controlling disulfide bond formation. The fact that immunoglobulin oxidation can be manipulated by a defined redox enzyme has implications for the control of humoral immunity.
Results
Both Ero1-Lα and Ero1-Lβ accelerate oxidative folding of a soluble protein in the ER
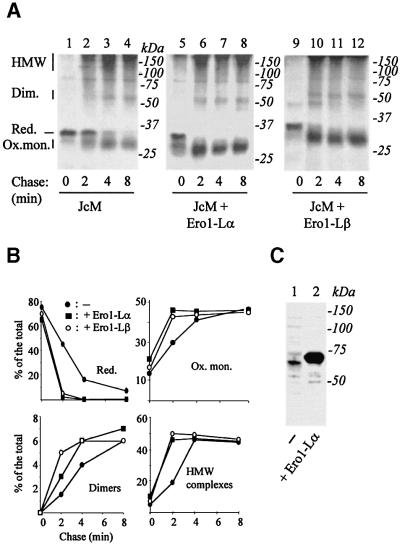
J chains are subunits of polymeric immunoglobulins (Ig) with three intra-chain disulfide bonds (Frutiger et al., 1992), the formation of which can be monitored by electrophoresis under non-reducing conditions (Mancini et al., 2000). Unassembled J chains are retained in the ER and eventually degraded by cytosolic proteasomes in both myeloma (Mancini et al., 2000) and HeLa cells (A.Mezghrani and R.Sitia, unpublished data). To facilitate serological detection, we tagged murine J chains with the myc epitope. To synchronize disulfide bond formation and to uncouple oxidation and isomerization, JcM expressing HeLa transfectants were pulsed in the presence of dithiothreitol (DTT) (Braakman et al., 1992a; Valetti and Sitia, 1994), and then chased in the absence of the reducing agent (Figure 1A). As expected, the majority of JcM present at the end of the pulse migrated with a mobility characteristic of reduced J chains (lane 1). After 2 min of chase, more oxidized monomers, dimers and higher molecular weight complexes appeared, progressively increasing during the chase.
Fig. 1. Ero1-Lα and Ero1-Lβ accelerate oxidative folding of JcM. HeLa cells were co-transfected with pcDNA.3.JcM and pcDNA3.1-ERO1-Lα (lanes 5–8), pcDNA3.1-ERO1-LβHA (lanes 9–12) or an empty vector (lanes 1–4) as a control. Forty-eight hours after transfection, cells were pulsed for 5 min with radioactive amino acids in the presence of DTT (3 mM), washed once at 4°C and chased for the indicated times without the reducing agent. (A) Anti-myc IPs were resolved under non-reducing conditions. The mobility of reduced JcM (Red.), oxidized monomers (Ox. mon.), covalent dimers (Dim.) and high molecular weight complexes (HMW) is indicated on the left-hand margin. (B) The different redox isoforms were quantified by densito metry, and plotted as the per cent of total JcM chains present at each chase time. JcM alone (filled circle); JcM + Ero1-Lα (filled square); JcM +Ero1-Lβ (empty circle). (C) Exogenous Ero1-Lαmyc is expressed at higher levels than endogenous Ero1-Lα. Western blot analysis with anti-Ero1-Lα (D5) from the lysates of 3 × 105 HeLa cells are shown for mock (lane 1) or pcDNA3.1-ERO1-Lαmyc (lane 2) transfected cells. Note the slower mobility of exogenous Ero1-Lα, caused by the presence of a C-terminal tag.
In HeLa cells over-expressing either Ero1-Lα or Ero1-Lβ, reduced J chains were no longer detectable already after 2 min of chase (lanes 6 and 10). In Ero1-Lα transfectants, more oxidized JcM monomers were visible already at the end of the pulse (lane 5). These results indicated that both Ero1-Lα and Ero1-Lβ accelerate the oxidative folding of a cargo protein in mammalian cells or make over-expressing cells more resistant to DTT. Densitometric quantification (Figure 1B) indicated that human hEROs accelerate the formation of both intra- and inter-chain disulfide bonds, without significantly altering the final equilibrium.
Western blot analyses with an anti-Ero1-Lα polyclonal antibody (D5) were performed to quantify the expression of exogenous Ero1-Lα, relative to endogenous molecules (Figure 1C). In different transfections, we measured a 10–15-fold higher signal in transfected cells than in control samples. Considering that ∼40% of cells were transfected, we estimated that individual cells expressed on average a >25-fold excess of the transgene. It should be noted, however, that the levels of expression varied considerably in different cells. Consistent with the notion that ERO1-Lβ is an UPR-induced gene (Pagani et al., 2000) we failed to detect endogenous Ero1-Lβ in non-stimulated HeLa cells. In most experiments, exogenous Ero1-Lβ was expressed at lower levels than exogenous Ero1-Lα (see Figure 4 for a comparison).
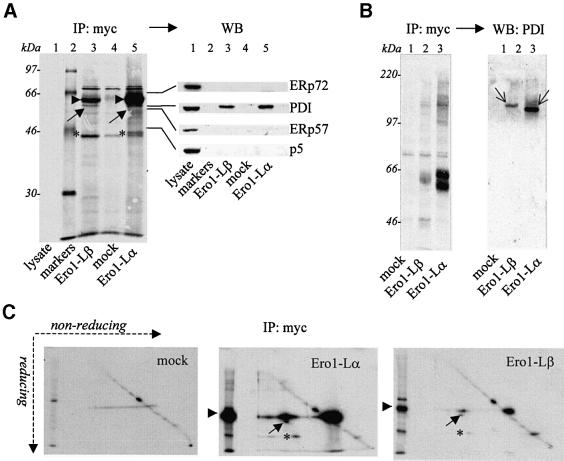
Fig. 4. Both Ero1-Lα and Ero1-Lβ form mixed disulfides with PDI. HeLa cells transfected with pcDNA3.1-ERO1-Lαmyc or pcDNA3.1-ERO1-Lβmyc were pulse-labelled for 15 h and immunoprecipitated with immobilized anti-myc. Blots were first exposed for autoradiography (IP: myc) and then decorated (western blot) with the indicated antibodies. IPs correspond to 1 × 106 cells per lane. (A) Gels were run under reducing conditions. Arrowheads point to Ero1-Lβ (lane 3), Ero1-Lα (lane 5), and the diagonal arrow to a 55 kDa band which is decorated by anti-PDI. In lane 1A, the total cell lysate from 2 × 105 unlabelled HeLa cells was run to provide positive controls for western blots. Lane 2 contains molecular weight markers. The asterisks (*) indicate a protein that interacts with both hEROs. (B) Gels were run under non-reducing conditions. Thin diagonal arrows point to the mixed disulfides between PDI and Ero1-Lβ (lane 2) or Ero1-Lα (lane 3). (C) Anti-myc IPs were resolved by ‘diagonal’ gels. A reduced sample for each condition was run in the second dimension (left lane). The spots indicated by arrows correspond to PDI. As in (A), the asterisks indicate a protein that interacts with both hEROs. Note that, like in most transient transfections, Ero1-Lβ was expressed at lower levels than Ero1-Lα (∼7-fold in the experiment shown).
During oxidative folding, JcM forms covalent complexes with PDI but not with Ero1-Lα or Ero1-Lβ
The effects of hEROs on J chain oxidative folding could either be due to a direct interaction between hEROs and JcM or be mediated by PDI and/or other ER oxidoreductases. To distinguish between these possibilities, the anti-myc immunoprecipitates (IPs) from pulse–chase assays were resolved under reducing conditions and blotted onto nitrocellulose (Figure 2A). The blot was first developed for autoradiography to follow radioactive JcM. The acceleration of oxidative folding in cells over-expressing hEROs was confirmed also under reducing conditions, as indicated by the appearance of a faster migrating JcM band (see left panels, Figure 2A). As demonstrated in Figure 2B, this electrophoretic shift is caused by the decrease in N-ethyl maleimide (NEM) molecules binding to oxidized J chains.
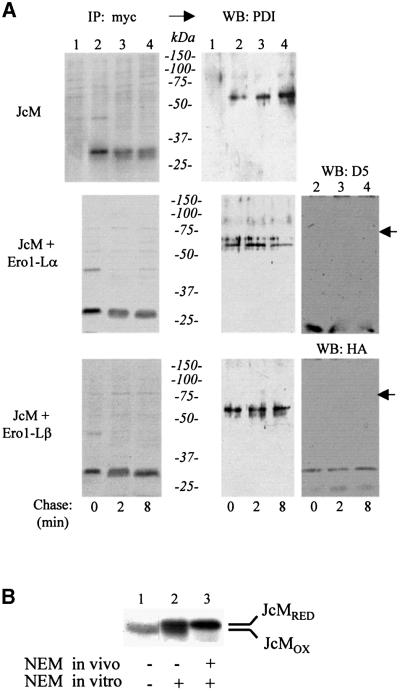
Fig. 2. JcM chains interact with PDI, but not with Ero1-Lα or Ero1-Lβ. (A) HeLa cells (2 × 106 cells per time point) expressing JcM alone or in combination with Ero1-Lα or Ero1-Lβ–HA, were pulsed and chased as described in legend to Figure 1 before lysis and immunoprecipitation with immobilized anti-myc antibodies. IPs were resolved by SDS–PAGE under reducing conditions and blotted onto nitrocellulose. Filters were first exposed and developed for autoradiography (IP: myc) and then analysed by western blotting (WB) with the indicated antibodies. Lane 1 shows the anti-myc IPs from mock-transfected cells labelled for 5 min. The arrows on the right hand margin point to the expected mobility of Ero1-Lα and Ero1-Lβ. (B) Oxidized and reduced JcM chains can be distinguished by differential binding to NEM. HeLa cells expressing JcM were pulse-labelled for 5 min in the absence of DTT and lysed in the presence (lanes 2 and 3) or absence (lane 1) of NEM, an alkylating agent, which binds only to reduced cysteines. Anti-J IPs were boiled for 5 min in sample buffer containing 50 mM DTT and resolved under reducing conditions. In lane 3, NEM (100 mM) was added before loading, so as to alkylate all JcM cysteines.
No bands corresponding to Ero1-Lα or Ero1-Lβ were detected associated to JcM when the same nitrocellulose filters were immuno-decorated with specific antibodies (D5 or anti-HA, respectively) (Figure 2A, right panels). In contrast, anti-PDI recognized a 55 kDa band in the anti-myc IPs from JcM transfectants (lanes 2–4), but not from mock-transfected HeLa cells (lane 1), indicating an interaction between JcM and PDI. PDI was detected at all chase times regardless of the presence of co-transfected hEROs, albeit at variable intensity.
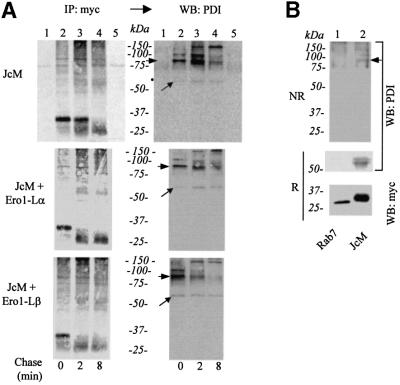
To determine whether the interaction between PDI and JcM was covalent, samples from a similar experiment were analysed under non-reducing conditions (Figure 3A). Several bands could be detected when blots were decorated with anti-PDI (lanes 2–4). These bands were absent from mock-transfected HeLa cells (lane 1) or when no anti-myc antibody was added during immunoprecipitation (lane 5). Whereas some free PDI could be detected (diagonal arrows), the majority of the signal originated from higher molecular weight disulfide-linked complexes. The molecular weight of the prominent band (80 kDa, arrowheads) was consistent with a JcM–PDI complex. A similar band was evident in the autoradiograms, indicating that it contained radioactive JcM chains. This band was recognized also by decorating the blot with anti-myc (our unpublished results) further confirming its identification. As the chase proceeded, the 80 kDa PDI–JcM mixed disulfide decreased in intensity, in correlation with the oxidation of JcM. Concomitantly, further bands of higher molecular weight appeared. The composition of these complexes, which contained at least a JcM and a PDI molecule, remains to be elucidated.
Fig. 3. During their oxidative folding, JcM chains form mixed disulfides with PDI. (A) HeLa transfectants were pulsed, chased and immunoprecipitated as in Figure 2A. IPs were resolved by SDS–PAGE under non-reducing conditions and blotted. Filters were first exposed and developed for autoradiography (IP: myc) and then decorated with anti-PDI (western blot: PDI). Lane 1 shows the anti-myc IPs from mock-transfected cells, while lane 5 shows the precipitate obtained with a class-matched irrelevant antibody from JcM-transfected cells pulse-labelled for 5 min. The diagonal arrows indicate monomeric PDI, while arrowheads point to the JcM–PDI mixed disulfides. (B) HeLa cells expressing Rab7-Myc (lane 1) or JcM (lane 2) were immuno precipitated with immobilized anti-myc antibodies as described before. IPs were resolved by SDS–PAGE under non-reducing (upper part, NR) or reducing (lower part, R) conditions and analysed by western blotting (WB) with the indicated antibodies. The arrow indicates the PDI–JcM disulfide complex.
JcM–PDI mixed disulfides were detectable also in cells that had not been treated with DTT (Figure 3B). In contrast, no PDI was co-precipitated by anti-myc from HeLa cells expressing a myc-tagged Rab7 (Figure 3B), further confirming the specificity of the assay. Taken together, these results identify mixed disulfides between PDI and a soluble cargo protein, JcM, during its oxidative folding in the ER.
hEROs form mixed disulfides with PDI
To identify proteins associated to Ero1-Lα or Ero1-Lβ, HeLa transfectants were metabolically labelled overnight and immunoprecipitated with anti-myc (Figure 4). Several bands co-immunopurified with Ero1-Lα and Ero1-Lβ were absent from mock-transfected cells (Figure 4A, compare lanes 3–5). To determine whether any of them contained one of the known ER oxidoreductases, the blot was sequentially probed with antibodies specific for ERp72, ERp57, PDI and P5 (western blot). The four antibodies decorated bands of comparable intensity in the lysates of HeLa cells (lane 1). However, only PDI was found associated to Ero1-Lα or Ero1-Lβ (lanes 5 and 3, respectively). Under non-reducing conditions (Figure 4B), both Ero1-Lα and Ero1-Lβ appeared to form both inter- and intra-chain disulfide bonds. As reported previously (Benham et al., 2000) two Ero1-Lα isoforms, OX1 and OX2, were clearly detectable. Anti-PDI decorated mainly bands of high molecular weight (see thin arrows), indicating that PDI bound covalently to Ero1-Lα or Ero1-Lβ.
To better visualize the patterns of covalent interactions of the two hEROs, we used diagonal, non-reducing/reducing gels (Figure 4C). In this system, proteins with intra-chain or inter-chain disulfide bonds run above or below the diagonal, respectively. In agreement with the results shown in Figure 4A and B, both Ero1-Lα and Ero1-Lβ covalently bound PDI (see arrow) mainly in binary complexes, and other unidentified cellular proteins, the most abundant of which yielded a band of ∼44 kDa (see asterisk) upon reduction. The electrophoretic pattern suggested that Ero1-Lα and Ero1-Lβ could also form covalent homodimers.
hEROs oxidize PDI but not ERp57
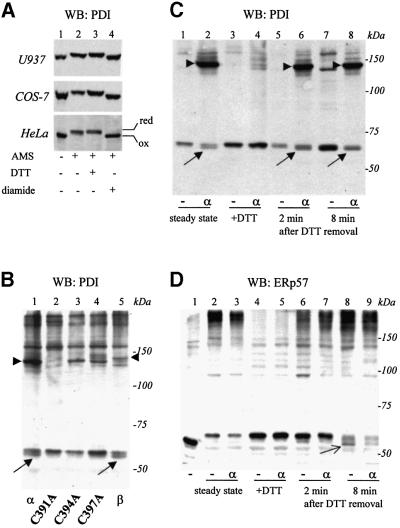
The alkylating agent 4′-maleimidylstilbene-2,2′-disulphonic acid (AMS) (Kobayashi et al., 1997), was used to analyse the PDI redox state in three different cell lines (U937, COS-7 and HeLa). The vast majority of PDI migrated more slowly after AMS modification (Figure 5A, compare lanes 1 and 2). Treatment of cells with the oxidant diamide prevented the mobility shift (lane 4) confirming that AMS reacted mostly with cysteine residues. In contrast, when cells were treated with DTT before lysis, the mobility of PDI was further decreased (lane 3). These findings indicated that PDI is partially reduced in mammalian cells and that its redox state can be manipulated by adding reducing or oxidizing agents (Bauskin et al., 1991; Braakman et al., 1992b).
Fig. 5. hEROs modulate the redox state of PDI but not of ERp57. (A) Redox state of PDI. U937, COS-7 or HeLa cells were incubated for 5 min at 37°C with DTT (lane 3), diamide (lane 4) or in medium alone (lanes 1 and 2) before lysis in TCA. Free protein thiols were modified with AMS (lanes 2–4) or not (lane 1) as described in Materials and methods and samples were resolved by SDS–PAGE (8% acrylamide). PDI was detected by western blotting. (B) hEROs modulate the redox state of PDI. HeLa cells over-expressing wild-type Ero1-Lα (α), Ero1-Lα cysteine replacement mutants (C391A, C394A, C397A), or Ero1-Lβ (β), were lysed in TCA. Precipitates were modified with AMS and processed as above. Diagonal arrows indicate oxidized PDI. hERO–PDI mixed disulfides are indicated by arrowheads. (C) Ero1-Lα promotes PDI re-oxidation. Mock (–) or Ero1-Lα (α) transfected HeLa cells were exposed to DTT for 5 min (lanes 3–8) and then cultured without the reducing agent (lanes 5–8). At the indicated times, cells were lysed in TCA and processed as above. Control samples with no DTT treatment are shown in lanes 1–2. Arrowheads point to the PDI–Ero1-Lα mixed disulfides, and diagonal arrows to oxidized PDI. (D) Ero1-Lα does not affect the ERp57 redox state. Mock or Ero1-Lα transfected HeLa cells were treated as in (C). The blot was decorated with anti-ERp57. A control sample with no AMS treatment is shown (lane 1); AMS treated samples with no DTT incubation are shown in lanes 2 and 3. Arrow indicates a band that likely corresponds to oxidized ERp57.
Unlike in untransfected HeLa cells, a faster migrating PDI band was detectable in cells over-expressing Ero1-Lα or Ero1-Lβ, (Figure 5B, lanes 1 and 5, respectively, diagonal arrows). Also, higher molecular weight bands became detectable. Amongst these, the bands indicated by arrowheads displayed mobility indistinguishable from the PDI-containing bands precipitated by anti-myc from Ero1-Lα or Ero1-Lβ transfected cells (see Figure 4B). Re-probing the blot with anti-myc decorated these bands (our unpublished results), confirming that they corresponded to mixed disulfides between PDI and Ero1-Lα or Ero1-Lβ. The Ero1-Lβ PDI mixed disulfide migrated slightly more slowly.
The detection of a faster migrating PDI band suggested that both Ero1-Lα and Ero1-Lβ could alter the redox state of this oxidoreductase. To further substantiate this finding, HeLa cells were transfected with Ero1-Lα cysteine-alanine replacement mutants in the conserved CXX CXXC motif. We showed previously that mutants in the second or third cysteines (C394A and C397A, respectively) are unable to complement the yeast ero1-1 strain (Cabibbo et al., 2000). Consistent with our previous findings (Benham et al., 2000), both mutants formed mixed disulfides with PDI (Figure 5B, lanes 3 and 4). However, they failed to induce the appearance of the faster migrating PDI band. Oxidized PDI also was not detected in HeLa cells expressing the Ero1-LαC391A, a mutant that partially complements ero1-1. Noteworthy, discrete PDI–C391A mixed disulfides were not detected in the 120 kDa region, but higher up in the gel, suggestive of larger oligomeric complexes.
To correlate the effects of hEROs on the oxidative folding of JcM chains and on the redox state of PDI, HeLa transfectants were treated with DTT for 5 min and then incubated for different times in the absence of the reducing agent. Each sample was treated with AMS and the electrophoretic mobility of PDI analysed under non-reducing conditions. Consistent with the results shown in Figure 5B, PDI was more oxidized in Ero1-Lα over-expressing cells (Figure 5C, compare lanes 1 and 2) and was reduced by exposing cells to 3 mM DTT (lanes 3 and 4). Already 2 min after DTT removal some oxidized PDI could be detected in Ero1-Lα over-expressing cells (lane 6, see arrow), but not in mock transfectants (lane 5). After 8 min, oxidized PDI was also detectable in mock-transfected cells. The abundance of oxidized PDI correlated with JcM oxidation in both mock and Ero1-Lα transfected cells (compare Figures 3 and 5).
As ERp57 has been shown to interact covalently with nascent proteins (Molinari and Helenius, 2000), we asked whether the redox state of this oxidoreductase was controlled by Ero1-Lα. As observed with PDI, treatment with AMS caused a pronounced mobility shift in ERp57 (Figure 5D, compare lanes 1 and 2), suggesting that also this oxidoreductase is found predominantly in the reduced state in HeLa cells. Interestingly, oxidized ERp57 isoforms (Figure 5D, lanes 8 and 9, see thin arrow) became detectable upon DTT recovery. However, neither at steady state nor during recovery from a reducing stress was the ERp57 redox state affected by over-expression of Ero1-Lα.
Cysteines in the CXXCXXC domain are essential for the function of Ero1-Lα
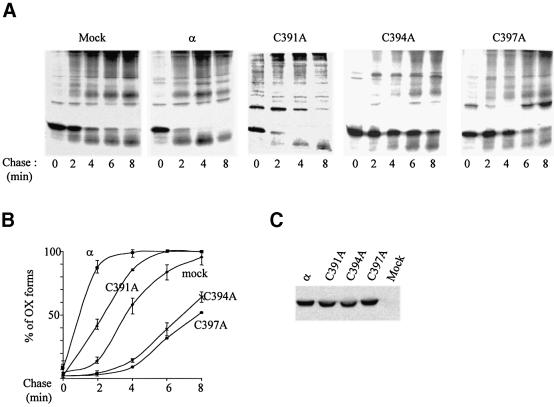
In cells over-expressing wild-type Ero1-Lα (Figures 1 and 6A), most JcM is oxidized after 2 min of chase. The effects of the CXXCXXC motif Ero1-Lα mutants on JcM oxidation were dramatically different (Figure 6A). C391A accelerated JcM oxidative folding, albeit less efficiently than the wild-type protein. In contrast, C394A and C397A inhibited oxidation. In C394A over-expressing cells, most JcM remained reduced even after 8 min of chase. Densitometric quantification from three independent experiments (Figure 6B) confirmed that C391 is important, but dispensable for the function of Ero1-Lα, whereas the replacement of C394 or C397 created dominant-negative forms.
Fig. 6. Ero1-Lα cysteine mutants inhibit JcM oxidative folding. HeLa cells expressing JcM were transfected with wild-type ERO1-Lαmyc (α) or myc tagged cysteine replacement mutants (C391A, C394A, C397A). (A) C394A and C397A act as dominant negatives. Pulse–chase experiments were performed as described in the legend to Figure 1A. Lysates were immunoprecipitated with anti-J. (B) The percentage of oxidized JcM was calculated by densitometry relative to the total JcM present at the end of the pulse. Average and standard deviations from two (C397A) or three (all other transfectants) experiments. (C) Mutants and wild-type Ero1-Lα are expressed at similar levels. Western blot analysis (anti-myc) from the lysates of 2 × 105 HeLa cells is shown.
hEROs control the oxidative folding of Ig-κ chains
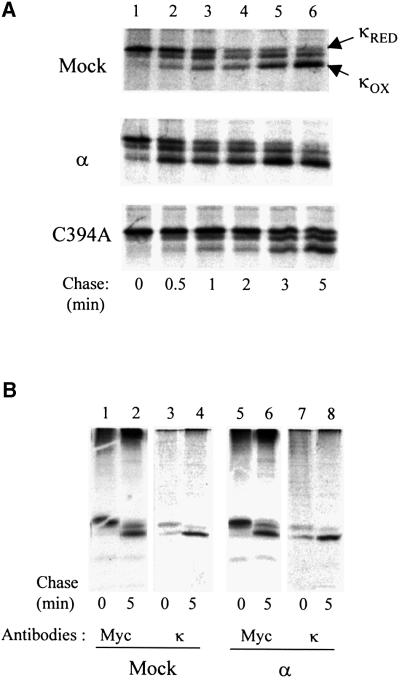
Having shown that hEROs control the oxidation rate of a terminally misfolded glycoprotein (JcM), we analysed the effects of wild-type Ero1-Lα (α) and C394A on Ig-κ chain, a non-glycosylated, secreted protein. As described for JcM, Ero1-Lα clearly accelerated the oxidation of myc-tagged κ chains upon DTT removal (Figure 7A). Even more dramatic were the effects of the dominant negative C394A mutant, which inhibited formation of intra-chain disulfide bonds in κ chains.
Fig. 7. The PDI–hEROs pathway is involved in the oxidative folding of Ig-κ chain, a non-glycosylated, secreted cargo protein. (A) HeLa cells were co-transfected with pcDNA.3-κ-myc and pcDNA3.1-ERO1-Lα or pcDNA3.1ERO1-LαC394Α. Cells were pulse-labelled for 5 min in the presence of DTT and chased without the reducing agent for the indicated times before lysis and immunoprecipitation with anti-myc. (B) Lysates from HeLa transfectants labelled as described in (A) were immunoprecipitated with anti-myc or with an antibody raised against native κ chains (goat anti-mouse κ) and resolved under non-reducing conditions. Note that anti-κ does not precipitate completely reduced κ chains.
Unlike anti-myc, antibodies raised against native κ chains did not recognize completely reduced molecules and only weakly immunoprecipitated κ chains in which only one of the two intra-chain bonds is formed (Figure 7B). In contrast, they quantitatively precipitated oxidized κ, suggesting that the formation of intra-chain disulfide bonds largely corresponds to productive folding (Hendershot et al., 1996).
Some high molecular weight material was precipitated by anti-myc but not by anti-κ (Figure 7B), suggesting that some κ chains aggregated following a DTT pulse. However, the over-expression of Ero1-Lα did not significantly increase the formation of covalent aggregates (Figure 7B).
Discussion
There is general agreement that the exocytic pathway is more oxidizing than the cytosol (Hwang et al., 1992; Cuozzo and Kaiser, 1999), but it is not entirely clear how this gradient is generated. In the absence of a direct means to measure the local ER redox conditions, we monitored disulfide bond formation in two soluble Ig subunits: J chains, which carry a single N-glycan and are unable to reach a transport competent structure (Mancini et al., 2000), and κ chains, which can be exported from the ER (Reddy et al., 1996; S.Ingrassia, A.Mezghrani and R.Sitia, unpublished results). Owing to the rapid formation of disulfide bonds in nascent Ig chains (Bergman and Kuehl, 1979) we used DTT to uncouple oxidation from translocation (Valetti and Sitia, 1994) and more easily uncover potential effects of hEROs. Using this technique, disulfide bond formation can be synchronized and, as importantly, a cysteine oxidation wave can be studied with little concomitant disulfide isomerization.
With this assay, we showed that both Ero1-Lα and Ero1-Lβ accelerate oxidative protein folding in the ER of human cells. This function is dependent on the presence of a functional hERO, as mutants in the conserved CXXCXXC motif show different activities. As in yeast complementation assays (Cabibbo et al., 2000), the C391A mutant displayed weaker activity on the oxidation of JcM, whereas the second and third cysteines were essential for proper function. The C394A and C397A mutants inhibited oxidative folding of both JcM and κ chains, possibly competing with endogenous Ero molecules for binding to PDI and/or to the recharging machinery. We cannot exclude a dominant-negative effect by direct interaction with endogenous Ero1-Lα. Inhibiting oxidative folding by introducing non-functional oxidoreductins could represent an important tool to identify the natural substrates of the hEROs–PDI driven pathway and analyse the cellular responses to redox imbalances.
Ero1-Lα and Ero1-Lβ accelerate the generation of both intra-chain and inter-chain bonds. The absence of mixed disulfides with JcM suggests that their activity is not due to direct interactions with cargo proteins. Thus, hEROs do not seem to have direct chaperone functions, but rather modulate the activity of PDI. Indeed, covalent complexes containing one PDI and one hERO are readily detected in HeLa transfectants (Figures 4 and 5). These complexes are present also in stable HeLa transfectants expressing low amounts of the tagged-transgene, suggesting that they are not artefacts due to over-expression, but rather intermediates in the electron transfer chain in the ER lumen. This interpretation is supported by the absence of such complexes from deletion or replacement Ero1-Lα mutants that show no functional activity (T.Simmen, G.Bertoli and R.Sitia, unpublished observations). The weaker activity of Ero1-LαC391A may depend on the formation of transient mixed disulfides with PDI (Benham et al., 2000) or on the larger complexes formed by this mutant. The presence of hEROs–PDI and PDI–JcM mixed disulfides but not of hEROs–JcM complexes 7suggests that electrons are transferred from nascent cargo proteins to hEROs via PDI (Figure 8). The mechanisms responsible for the re-oxidation of hEROs remain to be established. Whereas covalent complexes with PDI are easily detectable, we did not detect mixed disulfides between hEROs and three ER other abundant oxidoreductases, ERp57, ERp72 or p5. It is possible that these intermediates are too transient to be detected. On the other hand, the data in Figure 5D indicate that in human cells the redox state of ERp57 is not modulated by hEROs. This differs from that observed in S.cerevisiae, where Ero1p seems to transfer oxidative equivalents to PDI, Mpd2 (Frand and Kaiser, 1999) and possibly also Mpd1 (Norgaard et al., 2001).
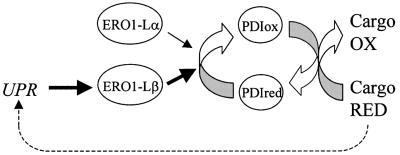
Fig. 8. Control of disulfide bond formation in mammalian cells. hEROs oxidize PDI, which in turn favours oxidation of cargo proteins. During ER stress, Ero1-Lβ is induced, while Ero1-Lα constitutively controls oxidative folding. When Ero1-Lα activity is insufficient, accumulating reduced cargo might induce synthesis of Ero1-Lβ.
How then is ERp57 oxidized? An indirect role of PDI is not so likely as the ERp57 redox state was not affected by the presence of abundant oxidized PDI in HeLa transfectants. Other proteins endowed with oxidase activity, such as the sulphydryl oxidases found in the secretory compartment (Hoober et al., 1996, 1999), might be in charge of controlling ERp57. With its interactors calnexin and calreticulin, ERp57 is thought to form a folding pathway that assists a class of glycoproteins (Oliver et al., 1997, 1999; Molinari and Helenius, 2000). Neither J nor κ chains, which carry a single or no glycan respectively, associate with calnexin or calreticulin during folding. Instead they utilize BiP as a chaperone and the hEROs–PDI system for disulfide bond formation. The thought of parallel folding pathways in the ER using different chaperones and folding enzymes, and displaying different substrate specificities, needs to be further explored.
While in S.cerevisiae PDI is present predominantly in the oxidized state (Frand and Kaiser, 1999; Tu et al., 2000), we found that the vast majority of PDI is partially reduced in HeLa, COS and U937 cells. Although scarce at steady state, a more oxidized form of PDI becomes detectable in HeLa cells soon during the recovery from DTT exposure, suggesting that cells rapidly activate robust responses to restore the optimal redox levels. As similar situation is observed for ERp57, it seems that the molecule(s) that control the ERp57 redox state are also activated by exposure to DTT.
The small pool of oxidized PDI in mammalian cells suggests that oxidative equivalents are rapidly transferred to cargo proteins, in agreement with the decrease of PDI–JcM mixed disulfides after DTT removal (Figure 3). Thus, electrons flow from nascent proteins to either Ero1-Lα or Ero1-Lβ, via PDI (Figure 8). The topological restrictions imposed by protein–protein interactions may enable the cell to localize and contain oxidation and reduction within the same cellular compartment (Tortorella et al., 1998).
Our studies reveal that Ero1-Lα and Ero1-Lβ are similar with respect to oxidoreductase binding and in assisting PDI oxidation. Why do humans and mice have two genes for a conserved function, fulfilled by a single essential gene in yeast? Intriguingly, Ero1-Lβ shows a higher degree of similarity to yeast Ero1p than Ero1-Lα in the presumed active sites (Pagani et al., 2000) raising the possibility that mammalian cells have evolved more subtle ways to control the ER redox state. An excess of oxidation may be toxic for cells, favouring protein aggregation in the ER and possibly activating signalling pathways. Pre liminary data suggest that Ero1-Lβ is more active than Ero1-Lα in sustaining oxidative folding. When the oxidative power of Ero1-Lα is overwhelmed, unfolded proteins will accumulate and rapidly induce synthesis of Ero1-Lβ via the unfolded protein response pathway (Figure 8).
Materials and methods
Antibodies
The D5 antiserum was raised in rabbits against non-reduced, reduced and denatured forms of an amylose resin-purified, MBP–Ero1-Lα fusion protein expressed in Escherichia coli. Rabbit anti-J was a generous gift from Michael Parkhouse (Pirbright Laboratories, UK), anti-ERp72 and anti-ERp57 from Maurizio Molinari (IRB, Bellinzona, Switzerland) and anti-P5 from Neil Bulleid (University of Manchester, UK). Rabbit anti-PDI was used as described previously (Benham et al., 2000). Affinity-purified goat antibodies to mouse Ig-κ chains were from Southern Biotechnologies (Birmingham, AL). The murine monoclonal antibodies 9E10 and 12CA5 were used to detect myc- and HA-tagged proteins, respectively. To obtain immobilized anti-myc, 9E10 ascites were bound to protein G–Sepharose, washed and cross-linked with dimethylpimelymidate (DMP; Sigma) for 30 min at room temperature. The reaction was blocked with 0.2 M ethanolamine, pH 8. Beads were washed and stored in phosphate-buffered saline (PBS) supplemented with NaN3.
Construction of plasmids for mammalian expression
A HindIII–BamHI fragment containing the entire coding sequence of murine J chain was excised from pJ6.5R1-βact, a kind gift of Dr Ron Corley (Boston University, MA), and subcloned into pcDNA3.1/Myc-His(–)A (Invitrogen). To generate JcM, PCR was performed with the forward primer 5′-AATACGACTACTATAG-3′ and the backward primer 5′-CAAGCTACCCGGGGTCAGGTAGC-3′ containing a SmaI site in the place of stop codon. With this protocol, an extra cysteine was inserted between the J coding region and the tag. This extra cysteine facilitated the detection of inter-chain disulfide bonds. Some covalent dimers and oligomers are seen also when untagged J chains are expressed in HeLa cells (our unpublished results). The amplified fragment was cloned HindIII–EcoRV in pcDNA3.1 in frame with the myc tag. Vectors encoding wild-type human Ero1-Lβ, Ero1-Lα and its mutants C391A, C394A, C397A have been described previously (Cabibbo et al., 2000; Pagani et al., 2000). To obtain ERO1-Lβ–HA, a PCR product containing the whole coding sequence (Pagani et al., 2000) was cloned BamHI–SmaI into the yeast expression vector pXYO12 in frame with the tag. The XbaI–Xho insert encoding for ERO1-Lβ–HA was subcloned in pcDNA3.1(–). Wild-type or mutant ERO1-Lα without the myc tag were used in co-transfections.
HeLa, COS-7 and U937 cells were obtained from ATCC. HeLa cells were transiently transfected by Lipofectin (Life Technologies) as described (Cabibbo et al., 2000) and analysed 40–48 h after transfection.
Metabolic labelling, immunoprecipitation, gel electrophoresis and western blotting
Western blot and pulse–chase analyses were performed as described (Mancini et al., 2000). For pulse–chase experiments, ∼5 × 106 transfected HeLa cells were used. To minimize disulfide bond rearrangements, NEM (10 mM final) was added to cells before lysis. Cells were lysed in RIPA buffer [0.2% SDS, 1% Nonidet P-40 (NP-40), 150 mM NaCl, 10 mM Tris–HCl, pH 7.6] with NEM (10 mM) for 30 min on ice. For long labelling, [5S]Promix (Amersham Italy, Milano, Italy) was added in complete medium (50 µCi/ml). After 15 h at 37°C, cells were lysed as described above.
Lysates were centrifuged at 10 000 g for 20 min to remove insoluble material, and incubated overnight with specific antibodies and protein A–Sepharose. Beads were resupended in buffer C (0.5% SDS, 1% NP-40, 150 mM NaCl, 10 mM Tris–HCl, pH 7.5) and loaded on a two-phase sucrose gradient (upper phase: 10% sucrose, 0.5% SDS, 1% NP-40, 150 mM NaCl, 10 mM Tris–HCl, pH 7.5; lower phase: 20% sucrose, 10 mM Tris–HCl, pH 7.5). Two additional washes were performed in buffers A (0.05% SDS, 1% NP-40, 500 mM NaCl, 10 mM Tris–HCl, pH 7.5) and B (0.05% SDS, 1% NP-40, 150 mM NaCl, 10 mM Tris–HCl, pH 7.5). IPs were resolved by standard or ‘diagonal’ SDS–PAGE (Reddy et al., 1996). Films were scanned by an automated densitometer (Molecular Dynamics, Sunnyvale, CA) and individual bands were quantified by the ImageQuant software.
Determination of the oxidation state of proteins
Cells were incubated for 5 min at 37°C with or without DTT (3 or 10 mM) or diamide (5 mM), and lysed immediately or after different incubation times in DTT-free medium at 37°C. To prevent post-lysis disulfide exchange, cells were directly precipitated in 10% trichloroacetic acid (TCA). Precipitates were washed in 70% acetone and resuspended in 40 µl of reaction buffer (80 mM Tris–HCl pH 6.8, 2% SDS), supplemented with a cocktail of protease inhibitors (Roche) with or without 25 mM freshly prepared AMS (Molecular Probes, Leiden, Netherlands). Samples were incubated for 30 min at room temperature and for 10 min at 37°C. A sample (10 µl) of non-reducing loading buffer was added and samples resolved by SDS–PAGE.
Acknowledgments
Acknowledgements
We thank N.Bulleid, A.Cabibbo and A.Helenius for helpful discussions, M.Molinari and R.M.E.Parkhouse for excellent antibodies, S.Ingrassia and A.Malgaroli for the myc-tagged Ig-κ vector, R.Corley for the J chain vector, C.Fagioli for help in the diagonal gels and S.Trinca for impeccable secretarial assistance. A.M. and T.S. are recipient of fellowships from EEC and Telethon, respectively. This work was in part supported through grants from AIRC, Consiglio Nazionale delle Ricerche (Target project on Biotechnology, PF49-97.0123 and 5% Biotechnology PF 31-00897), and Ministero della Sanità (RF 9853) to R.S., and from NWO-CW to I.B.
References
- Bauskin A.R., Alkalay,I. and Ben-Neriah,Y. (1991) Redox regulation of a protein tyrosine kinase in the endoplasmic reticulum. Cell, 66, 685–696. [DOI] [PubMed] [Google Scholar]
- Benham A.M., Cabibbo,A., Fassio,A., Bulleid,N., Sitia,R. and Braakman,I. (2000) The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lα. EMBO J., 19, 4493–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman L.W. and Kuehl,W.M. (1979) Formation of an intrachain disulfide bond on nascent immunoglobulin light chains. J. Biol. Chem., 254, 8869–8876. [PubMed] [Google Scholar]
- Braakman I., Helenius,J. and Helenius,A. (1992a) Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J., 11, 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Helenius,J. and Helenius,A. (1992b) Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature, 356, 260–262. [DOI] [PubMed] [Google Scholar]
- Cabibbo A., Pagani,M., Fabbri,M., Rocchi,M., Farmery,M.R., Bulleid,N.J. and Sitia,R. (2000) ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem., 275, 4827–4833. [DOI] [PubMed] [Google Scholar]
- Cuozzo J.W. and Kaiser,C.A. (1999) Competition between glutathione and protein thiols for disulphide-bond formation. Nature Cell. Biol., 1, 130–135. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Molinari,M. and Helenius,A. (1999) Setting the standards: quality control in the secretory pathway. Science, 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Frand A.R. and Kaiser,C.A. (1999) Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell., 4, 469–477. [DOI] [PubMed] [Google Scholar]
- Frutiger S., Hughes,G.J., Paquet,N., Luthy,R. and Jaton,J.C. (1992) Disulfide bond assignment in human J chain and its covalent pairing with immunoglobulin M. Biochemistry, 31, 12643–12647. [DOI] [PubMed] [Google Scholar]
- Gething M.J. and Sambrook,J. (1992) Protein folding in the cell. Nature, 355, 33–45. [DOI] [PubMed] [Google Scholar]
- Helenius A., Marquardt,T. and Braakman,I. (1992) The endoplasmic reticulum as a protein folding compartment. Trends Cell Biol., 2, 227–231. [DOI] [PubMed] [Google Scholar]
- Hendershot L.M., Wei, J, Gaut,J., Melnick,J., Aviel,S. and Argon,Y. (1996) Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc. Natl Acad. Sci. USA, 93, 5269–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober K.L., Joneja,B., White,H.B. and Thorpe,C. (1996) A sulphydryl oxidase from chicken egg white. J. Biol. Chem., 271, 30510–30516. [DOI] [PubMed] [Google Scholar]
- Hoober K.L., Sheasley,S.L., Gilbert,H.F. and Thorpe,C. (1999) Sulfhydryl oxidase from egg white. A facile catalyst for disulfide bond formation in proteins and peptides. J. Biol. Chem., 274, 22147–22150. [DOI] [PubMed] [Google Scholar]
- Hwang C., Sinskey,A.J. and Lodish,H.F. (1992) Oxidized redox state of glutathione in the endoplasmic reticulum. Science, 257, 1496–1502. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Kishigami,S., Sone,M., Inokuchi,H., Mogi,T. and Ito,K. (1997) Respiratory chain is required to maintain oxidized states of the DsbA–DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl Acad. Sci. USA, 94, 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini R., Fagioli,C., Fra,A.M., Maggioni,C. and Sitia,R. (2000) Degradation of unassembled soluble Ig subunits by cytosolic proteasomes: evidence that retrotranslocation and degradation are coupled events. FASEB J., 14, 769–778. [DOI] [PubMed] [Google Scholar]
- Molinari M. and Helenius,A. (1999) Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature, 402, 90–93. [DOI] [PubMed] [Google Scholar]
- Molinari M. and Helenius,A. (2000) Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science, 288, 331–333. [DOI] [PubMed] [Google Scholar]
- Norgaard P., Westphal,V., Tachibana,C., Alsoe,L., Holst,B. and Winther,J.R. (2001) Functional differences in yeast protein disulfide isomerases. J. Cell Biol., 152, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.D., van der Wal,F.J., Bulleid,N.J. and High,S. (1997) Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science, 275, 86–88. [DOI] [PubMed] [Google Scholar]
- Oliver J.D., Roderick,H.L., Llewellyn,D.H. and High,S. (1999) ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol. Biol. Cell., 10, 2573–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M., Fabbri,M., Benedetti,C., Fassio,A., Pilati,S., Bulleid,N.J., Cabibbo,A. and Sitia,R. (2000) Endoplasmic reticulum oxidoreductin 1-lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J. Biol. Chem., 275, 23685–23692. [DOI] [PubMed] [Google Scholar]
- Pollard M.G., Travers,K.J. and Weissman,J.S. (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell., 1, 171–182. [DOI] [PubMed] [Google Scholar]
- Reddy P., Sparvoli,A., Fagioli,C., Fassina,G. and Sitia,R. (1996) Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled Ig light chains. EMBO J., 15, 2077–2085. [PMC free article] [PubMed] [Google Scholar]
- Tortorella D., Craig M.S., Huppa,J.B, Wiertz J.H.J., Jones T.R., Ploegh H.L. (1998) Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol., 142, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B.P., Ho-Schleyer,S.C., Travers,K.J. and Weissman,J.S. (2000) Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science, 290, 1571–1574. [DOI] [PubMed] [Google Scholar]
- Valetti C. and Sitia,R. (1994) The differential effects of dithiothreitol and 2-mercaptoethanol on the secretion of partially and completely assembled immunoglobulins suggest that thiol-mediated retention does not take place in or beyond the Golgi. Mol. Biol. Cell., 5, 1311–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]