Abstract
Background and objective:
Endophytic fungi, particularly Trichoderma species, offer significant potential for the isolation of novel natural bioactive compounds with diverse applications in agriculture, medicine, and the food industry. These compounds are reliable, cost-effective, and environmentally safe. In this study, we evaluated the efficacy of secondary metabolites extracted from Trichoderma species against the JURKAT cell line.
Methods:
Trichoderma species were utilized in the fermentation process to produce secondary metabolites, which were subsequently evaluated for anticancer activity against the JURKAT cell line using MTT, LDH, apoptotic assays, and gene expression studies.
Results:
The study demonstrated that the ethyl acetate extract of Trichoderma secondary metabolites significantly reduced cell viability and increased LDH leakage, indicating potent anti-proliferative activity against the JURKAT cell line. Cytotoxicity ranged from 35.12 ± 0.45% to 58.56 ± 1.16% at varying concentrations. The extract significantly increased the percentage of cells arrested in the G2/M phase from 8.48% to 21.11%, while reducing cell viability from 94.3% to 54.35% and elevating early apoptotic cells from 1.12% to 21.34%. Real-time PCR analysis revealed a dose-dependent upregulation of the caspase 3 gene, whereas caspase 8 was downregulated.
Conclusion:
The secondary metabolites extracted from Trichoderma show promise for further early-stage clinical trials, including pharmacodynamic studies and in vivo investigations using animal models. In conclusion, these extracts exhibit significant anti-carcinogenic potential and could serve as a therapeutic agent for treating leukaemia.
Key Words: Endophytes, Trichoderma, Apoptosis, Gene expression, Acridine orange
Introduction
A promising strategy for drug development is the ongoing search for secondary metabolites from microbes residing in unexplored habitats [1]. The chemical variety and medicinal potential of secondary metabolites generated from fungi make them attractive. In the fields of medicinal chemistry and drug development, endophytic fungi are continuously mined for potential pharmaceutical prospects. Endophytic fungal symbionts found in marine flora, terrestrial plants, and wildlife often generate intriguing secondary metabolites with biomedically significant antiviral, antituberculosis, and anticancer effects [2]. One of the primary origins of bioactive compounds employed in various aspects of medical treatment, including as cancer therapy, is thought to be endophytes. Once colonized, they either enhance the host plant’s machinery to create these beneficial compounds or produce them as part of their secondary metabolite production [3]. Thus, in recent decades, the scientific community has shown a heightened interest in the investigation of endophytes. Endophytic fungus makes up a significant fraction of the endophytic microbiota among the endophytes. Alkaloids, lignans, terpenes, polyketides, polyphenols, quinones, xanthenes, tetralones, peptides, and spirobisnaphthalenes are among the anti-cancer substances generated from endophytic fungi that have been shown to be protective against cancer [4]. Acremonium, Alternaria, Aspergillus, Ceriporia, Chaetomium, Colletotrichum, Cytospora, Emericella, Eurotium, Eutypella, Fusarium, Guignardia, Hypocrea, Penicillium, Pestalotiopsis, Phomposis, Periconia, Stemphylium, Talaromyces, Thielavia, Trichoderma and Xylaria are the endophytic fungal genera that contain two or more putative producers of anticancer agents. There is an alternate supply of bioactive chemicals provided by this endophytic fungus. Biotechnology and genetic engineering may help us improve their yield of particular anticancer drugs [5].
There is an increasing incidence of cancer cases year, which positions it as a prominent contributor to global mortality [6]. The increased death rate makes it imperative that researchers worldwide look for novel sources of anticancer medications. The determination and advancement of novel and enhanced chemotherapeutics derived from synthetic or natural sources has been a recent breakthrough in the treatment of cancer. The possibility of discovering novel structural classes with distinctive bioactivities for cancer treatment exists in natural sources. Bioactive metabolites from endophytic fungus are abundant and can be synthesized into novel and useful analogues for chemotherapy [7]. Compounds obtained from endophytic fungus have been scientifically demonstrated to be reliable and plentiful reservoirs of anticancer substances, which could have a substantial impact on the advancement of anticancer drugs in contemporary medicine [8]. The development of powerful medications with low side effects has provided a modern alternative to traditional approaches to illness prevention and treatment [9]. It was estimated in 2000 that natural products and their derivatives made up around 57% of the chemicals being tested in clinical trials for cancer therapy [10]. According to preliminary findings, endophyte-derived natural bioactive chemicals could be a potential alternative source for developing novel anticancer medications [11]. Antiproliferative tests are commonly used in studies on the cytotoxicity of metabolites from endophytic fungi. These tests involve evaluating many compounds simultaneously to determine their potential anticancer activity. Additionally, high-throughput screening is employed to efficiently assess the effects of these compounds on cancer cell lines [12]. Finding potential drugs can be accomplished by isolating naturally occurring bioactive chemicals and evaluating them for pharmacological characteristics [13]. It has been reported that a number of endophytic fungal strains generate novel chemicals that show promise in anticancer assays [14]. Several plant-derived anticancer medications, including vincristine, etoposide, irinotecan, topotecan, and vinblastine, are being used clinically to treat a variety of malignancies in humans [15]. The anti-cancer properties of Pongamia pinnata were found to be related to the targets (epidermal growth factors) EGFR, ERBB2, and EGF. According to Karamala et al. [16] in-silico research from 2024, the maximum binding affinities for the docked complexes were -9.5kcal/mol for ERBB2 with Vitexin, -8.2kcal/mol for EGF with Pazopanib, and -8.3 kcal/mol for EGFR with pongachromene. HPLC’s in-vitro investigations revealed a high concentration of flavonoids and phenols, which help prevent skin cancer. Thirty chemicals were identified by GC-MS analysis [17]. Higher amounts of fatty acids were found, whereas very little of other chemicals were separated. Dibutylphthalate (C16H22O4) has the highest concentration of the component, with a peak area of 23.87% observed with RT 18.49. Oleicacid (C18H34O2) has a peak area of 19.35 with RT 21.92, 1-Docosene (C22H44) has a peak area of 18.83, and oxalic acid, allyl hexa decyl ester (C21H38O4) has a peak area of 10.77. Cyclohexane, 1,1’-(1,2-dimethyl 1,2-ethanediyl) bis-(C16H30), Cyclohexane, hexyl- (C12H24), Decane2,3,5,8-tetramethyl (C14H30), and 2-Hexyl-1-octanol (C14H30O) are the remaining compounds. Dibutylphthalate (C16H22O4) was found to be anticancerous in nature by many studies [18]. The abundance of cultivable bacterial endophytes that exist in Pongamia pinnata bark was examined in detail in this work, along with the possible anticancer effects of their bioactive secondary metabolites. The anticancer ability of the secondary metabolites was evaluated by LDH, MTT, apoptotis, flow cytometry, acridine orange and gene expression studies by real-time PCR.
Materials and Methods
Materials and Reagents
Pongamia pinnata bark was obtained from the local stores, and the chemicals, reagents were obtained from Sigma-Aldrich, India: copper sulphate, (FBS) fetal bovine serum, trypsin, antibiotics (penicillin/streptomycin) and RPMI-1640 medium. The JURKAT cell line, which is a human acute T-cell leukemic cell line, was acquired from the American Type Culture Collection (ATCC) with the accession number TIB-152.
Collection and Processing of Samples
Bark was peeled off from the tree samples, identified, and certified from the Botanical garden campus, Bangalore. Specimens were obtained and placed in aseptic polythene bags, then transported to the laboratory to facilitate fungal cultivation. In order to isolate the fungus, the specimens undergo surface sterilization for 1 minute using a 1% solution of sodium hypochlorite (NaClO), followed by three consecutive rinses with distilled water, each lasting 1 minute. The uppermost layers of bark were subsequently removed using sterile forceps, and each sample, approximately 2 cm in size, was placed onto the surface of PDA (with 1gm streptomycin 2 mg/ml). The Petri plates were incubated at 27°C for a total of 15 days. Throughout this period, the dishes were regularly examined to monitor the development of any fungal growth. A small sample from each fungal species was collected and deposited on Potato Dextrose Agar (PDA) under sterile conditions to purify and identify the fungi.
Characterization and Identification of Fungal Endophytes
The fungal endophytes that were grown were identified by observing their morphology while growing on SDA. In order to enhance visibility of the hyphae, conidia, and their conidiophores when observed with a microscope, they were subjected to staining with lactophenol blue.
Molecular-Based Identification
Internal transcribed spacer (ITS)-based rDNA sequencing, as previously reported by Tibpromma et al. [19], was ultimately used to identify fungal endophytic strains, albeit with a few minor adjustments. Genomic DNA was extracted by placing about 50mg of fungal mat in liquid nitrogen and crushing them into powder. Following vortexing, the contents were placed in 1ml of cetyltrimethylammonium bromide (CTAB) buffer and incubated at 60°C for one hour. The contents were obtained by extracting using a mixture of 700 µL of phenol, chloroform, and isoamyl alcohol (in a ratio of 25:24:1) after centrifuging at 10,000rpm for 10 minutes. The upper aqueous layer was collected and the DNA was precipitated by adding 800µl of ice-cold isopropanol. Next, the tubes were gently turned upside down to thoroughly mix the contents, and then spun at a speed of 10,000rpm for 10 minutes at a temperature of 4°C. Subsequently, the pellets were purified using 70% ethanol and let to dry naturally. The DNA was reconstituted using 80µL of TE buffer and kept at -20°C. The DNA’s purity was assessed using a UV VIS spectrophotometer (Shimadzu, 1800). ITS-based primers were used [ITS1: 5′-TCCGTAGGTGAACCTGCGG-3′; ITS4: (5′TCCTCCGCTTATTGATATGC-3′] were used in amplifying 500–600bp of the genomic DNA extracted [20]. A total of 50μl of DNA (50–500ng) was used to set up the PCR reaction with both forward and reverse primers at 10nm concentration. PCR was allowed for 40 cycles with initial denaturation at 95°C for 5min, followed by 30sec at 94°C, 1min at 58°C, 1min at 72°C and a final extension of 12min at 72°C. The final PCR product was sequenced using an ABI 3730 sequencer (Applied Bio-systems, USA) after being extracted from an agarose gel using a HiMedia DNA purification Kit (HiMedia, India). The Basic Local Alignment Search Tool (BLAST) was used to evaluate the sequences. Phylogenetic analysis was done with MEGA-5.0 (neighbor-joining method).
Fermentation and Extraction of Secondary Metabolites
The isolated fungi was cultivated in Sabaroud Dextrose Broth (SDB) medium and incubated for 20days at 27°C with constant shaking at 200 rpm in order to extract the molecules. Following fermentation, the broth was initially filtered with a muslin cloth and then with a filter paper. The contents were then centrifuged for 10 min at 12,000 rpm. The clear supernatant was collected in a 250 ml separating funnel and was added with equal volumes of ethyl acetate (EA). The contents were kept for vigorous shaking on a shaker at room temperature for 24 hr. The upper organic layer was transferred to a fresh centrifuge tube and concentrated using a rotating vacuum evaporator. The resultant extract was subsequently diluted in a 10% solution of dimethyl sulphoxide (DMSO, 1mg/mL) and used for further analysis.
Phytochemical Analyses
Polyphenols, flavonoids, and tannins were examined using phytochemical analysis in ethyl acetate extracts derived from fermented samples of Trichoderma alone. Every test was conducted in three duplicates [21].
Estimation of Polyphenols
Folin-Ciocalteu reagent was employed for the spectrophotometric measurement of total polyphenols utilizing the colorimetric approach. To put it briefly, 200µL of extract and 800µL of sodium carbonate, along with 1ml of Folin-Ciocalteu reagent (1:10) were added to a test tube and incubated for thirty minutes, and OD was recorded at 765nm. Gallic acid (1mg/ml) was used as a reference to quantify the phenols.
Estimation of Flavonoids
Flavonoids were estimated as described by Kim et al. [22] but with slight modifications. In brief, 120μl of NaNO2 (5%) was mixed with 0.4ml of extract and mixed thoroughly. To the contents, 0.12ml of AlCl3 (10%) was added after 5min and mixed. After 6min of incubation, 800 μL of NaOH (1M) was added and OD was recorded at 510 nm. Quercetin (1mg/ml) was used as standard to quantify the flavonoids content.
Estimation of Tannis Content
The condensed tannins were estimated by the vanillin method. To 50μl of extract, 1.5 mL of the vanillin/methanol (1%) solution was added and mixed thoroughly. To the contents, 750 μl of hydrochloric acid was added and incubated at room temperature for 20 min. Following incubation, the OD was recorded at 550nm. Tannic acid (1mg/ml) was used as a reference standard to quantify the tannins content.
Cell culture and MTT assay
The RPMI-1640 media was added with 10% FBS along with penicillin and streptomycin to maintain the human acute T-cell leukemic cell lines (JURKAT). Cell viability was screened with 3-(4, 5-dimethylthiazole)-2,5-diphenyltetrazolium bromide (MTT) assay as described by Beheshti, F et al (2021) but with slight modifications. In this experiment, RPMI-1640 media was used to seed approximately 5x104 cells onto 96-well plates, which were then placed in a CO2 incubator. After a 24-hour incubation period, 100µL of new medium was added to each well, along with 10µL of MTT reagent (0.5mg/ml), and the mixture was left to incubate for 4hr. Different amounts of ethyl acetate extract was added as treatment to the cells (3.125, 6.25, 12.5, 25, 50 and 100 µg/ml). Positive control was vincristine (25 µM). After the wells were incubated, the solution was removed and OD was then measured at 590nm using a plate reader (Genetix, India) after adding 100µL of dimethyl sulphoxide (DMSO).
LDH Assay for Testing Cell Cytotoxicity
Using an LDH reagent (HiMedia, India), the LDH that leaked from the membrane disruption was calculated [23]. The enzyme released in this assay transforms into a red formazan product known as iodonitrotetrazolium violet (INT), a tetrazolium salt. The number of lysed cells is strongly correlated with the amount of colour that forms. Similar to the preceding section, cells were cultivated with different concentrations of extracts (10, 20, 40, 80, 160, and 320 µg/ml). Positive control was vinblastine (25 µM). After incubating for 24 hours, the medium was changed out for 50µL of fresh medium and 50 µL of LDH reagent (0.5 mg/ml), and then incubated for 30min in the dark. After incubation, the plate reader (Genetix) was used in measuring the absorbance at 490 nm. The following equation was employed for calculating the cytotoxicity
Cytotoxicity (%) = LDH release from the experiment / Maximum release of LDH.
Background sounds from the culture media were eliminated. The maximal amount of LDH released by these cells was 100%. Using SPSS (Faculty version), the half inhibitory doses (IC50) of green produced CuNPs were ascertained.
Cell Cycle Study on Jurkat Cells
In a 6-well plate with about 2 mL of serum-free medium, about 1x106 cells were planted and grown for 24 hr. Following treatment, cells were cultured for 24 hours at the appropriate concentrations of the supplied samples that had been prepared in full medium [24]. After the cells were extracted, they were spinned at 2000 rpm for 5 min, and the pellet obtained was washed with 2 ml of 1XPBS twice. Followed by washing, the pellet was then resuspended and added with 1 mL of cooled 70% ethanol drop by drop. Following incubation overnight at 4°C, the cells were centrifuged for 5 min at 2000 rpm after fixation. The pellet was washed thrice with 2 ml of ice-cold 1X PBS, and following a 15-minute dark incubation period, the pellet was again suspended in sheath fluid (Propidium Iodide (PI) and 0.05 mg/ml RNaseA).
With FACS Caliber (BD Biosciences) flow cytometry, the proportion of cells in each cell cycle stage was calculated for populations treated and untreated with chemicals [25].
Apoptosis Detection
Apopotosis was carried out according to Xia et al. [23] but with slight modifications. In brief, using cell culture media, 1X106 cells per well were plated on a 6-well plate the day before apoptosis was induced. The wells were examined for floating (dead) cells after 18 hours of incubation and discarded if any, maintaining the original volume with fresh culture medium [24]. After incubating for 24 hours, the cells were exposed to 40 and 80 µg/ml of the sample to trigger apoptosis. After that, the cell culture medium and the cells were taken off of the dish using a policeman and added to a centrifuge tube. The pellet obtained was washed with PBS twice, and the cells (1 x 106cells/ml) were resuspended in 1 ml of 1X Binding Buffer. To 500µL of the cell suspension, 10µL of PI and 5μL of Annexin V were added. The suspension was incubated at room temperature for fifteen minutes. Following incubation, the cells were examined using a flow cytometer produced by FCS Express software (FACS Caliber, BD Biosciences) in less than an hour.
Acridine Orange and Ethidium Bromide Staining
Approximately 25 microliters (1x105 cells per milliliter) of treated and untreated cells were placed in separate microcentrifuge tubes. Then, 5 microliters of AO-EtBr (Acridine orange and Ethidium Bromide) were added to each tube and allowed to stain the cells for approximately 2 minutes. 10µl of the cell suspension was applied onto a microscopic slide with a glass coverslip and observed using a fluorescence microscope (Compound microscope, Olympus, Japan) equipped with a fluorescein filter. The nuclear morphology should be clearly observable [26].
Gene Expression Studies
RNA Isolation
RNA extraction was done as described by Livak and Schmittgen, [27] but with slight changing; the cells were washed twice with ice-cold PBS. Then the Jurkat cells were adherently treated with about 2 mL of TRIzol and transferred to a fresh tube. Samples were left to stand at room temperature for 5 min, and a solution of 0.2 ml chloroform to 1 ml TRIzol was added. After sealing the tubes, they were vortexed vigorously and incubated at room temperature for five minutes. The resultant mixture was centrifuged for 15 min at 4 0C at 10,000 rpm. The upper aqueous phase was moved to a fresh tube containing 0.5ml of isopropanol and 1ml of TRIzol. Contents were incubated for five minutes and centrifuged for 10 minutes at 4 0C at 10,000rpm. The RNA pellet obtained was resuspended in 1ml of 70% ethanol and centrifuged again for 5min at 4 0C and 14,000 rpm. The pellet was dried using air and then reconstituted in 25 µl of DEPC-treated water [28].
RT-PCR
RT PCR (RT-PCR - CFX96 Biorad, USA) was performed somewhat differently from Schmittgen and Livak [26]. Following the manufacturer’s recommendations, 2µg of RNA were used to synthesize cDNA utilizing the Prime script RT reagent kit with oligodT and random hexamer primer. The mixture volume was adjusted to 20μl, and the cDNA synthesis was carried out for 30min at 52°C then inactivated at ambient temperature for 5min at 85°C and finally, the cDNA was diluted with twice the volume (1:2). The following reagent volumes are utilized in RT-PCR conditions: 0.5μl of cDNA, 0.3 μl of forward primer, 0.3μl of reverse primer, 10.0 μl of Sybr green, 0.4 μl of DMSO and 6.5 μl of nuclease-free water. The Comparative CT Method (ΔΔCt Method) is used to determine the relative fold expression of the target gene [29]. The CT method was used to measure the target gene’s relative expression in relation to the housekeeping gene (GAPDH) and untreated control cells. ΔΔCt is equal to ΔCt (control group) – ΔCt (treatment group). The formula used to compute the fold change in target gene expression for each treatment was fold change = 2(-ΔΔCt).
Statistical Analysis
The research was carried out with three sets of experiments, and the results are shown as mean ± SD. Student’s t-test was used to compare the experimental outcomes. A statistically significant P value was defined as one that was less than 0.05, or p<0.05.
Results
Isolation of Fungi
Two different forms of fungi were recovered from the SBD plates, out of which we found Trichoderma species to be predominant and with a higher frequency of appearance (0.81) than Colletotrichum species (0.19). Other species like Fusarium sp. and Alternaria sp. were found, but in very trace frequencies. We have only talked about the two types of fungi that we found in our study Trichoderma and Colletotrichum sp (Figure 1).
Figure 1.
Fungal Plates and Slides as Isolated in Our Study from the Sample. T, Trichoderma sp; C, Colletotrichum sp
The lactophenol blue-stained fungal hyphae have scattered conidiophores and appear slender. The sequences from the internal transcribed spacer (ITS) regions were used to identify the fungal strains as Colletotrichum brevisporium and Trichoderma viriens. Trichoderma sample sequence showed 97.79% identity to T. viriens Accession number NR_138428, and Colletotrichum sample showed 97.75% identity to C. brevisporium with Accession number PP905057.1. Phylogenetic tree construction was done using MEGA 7.
Phytochemical Analysis
Extraction was done with both Trichoderma and Colletotrichum samples. From our study, we found, with Trichoderma and Colletotrichum yield was found to be 2.31 and 0.96%. From the phytochemical analysis we found high tannin content of 1214.28 mg TAE (Tannic acid equivalent)/g DE for ethyl acetate extract followed by flavonoids of 564.78 mg QE (Quercetin equivalent)/g. Phenols were found to be 64.12 mg of Gallic acid equivalent per gm of ethyl acetate extract.
Evaluation of Extracts on JURKAT Cell Line
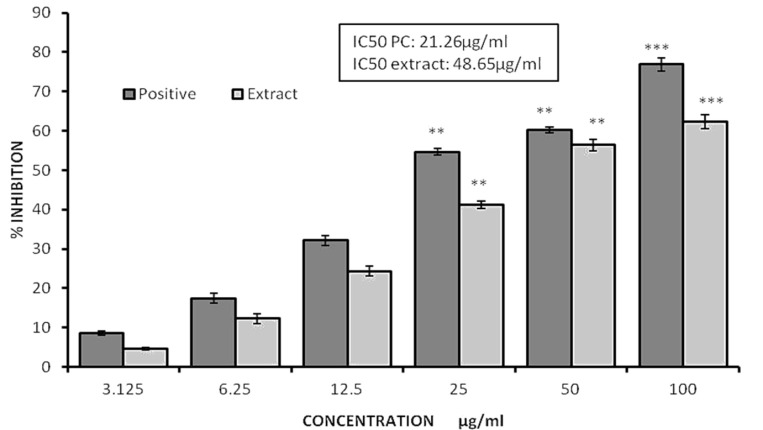
A mitochondrial toxicity test was conducted using the MTT reagent (Figure 2). The extract and positive control significantly decreased the cell viability in the investigation at concentrations of 21.26 µg/ml and 48.65 µg/ml, respectively. The ethyl acetate extract exhibited inhibition ranging from 41.23 to 62.34 at concentrations of 25 and 100µg/ml, respectively. In contrast, the positive control demonstrated inhibition of the cell lines ranging from 54.67 to 76.85 at concentrations of 25 and 100µg/ml respectively. In our investigation, we noticed that the extract exhibited a decrease in viability, as determined by the MTT assay, with an IC50 value of 48.65µg/ml. This reduction in viability was highly significant (p<0.05) when compared to control, demonstrating that the extract has anti-proliferative action. Conversely, the positive control showed an IC50 value of 21.26 µg/ml.
Figure 2.
Graphs Illustrating the Percentage of Inhibition of the Ethyl Acetate Extract against the JURKAT Cell Line as Determined by the MTT Experiment. All the values are average of triplicates and presented as value ±SD. ** P<0.01, *** P<0.001
LDH Assay
Our results from the MTT assay were validated by the LDH assay (Figure 3). The measurement of total cytotoxicity was a comparison between the levels of LDH released by extract and the cellular LDH as seen with positive control. A dose-dependent increase in LDH leakage was observed with our extract. At 25 and 1000 µg/ml, the JURKAT lines’ cytotoxicity ranged from 35.12 ± 0.45 to 58.56 ± 1.16%.
Figure 3.
Graphs Illustrating the Percent LDH Release of Extract against JURKAT Cell Line as Calculated with LDH assay. All the values are mean expressed as value ±SD. ** P<0.01, *** P<0.001
Cell Cycle Studies on JURKAT Cells
As mentioned previously, the extract’s IC50 value was 48.65 μg/ml. The anticancer activity was scheduled at 50 and 100 μg/ml concentrations, or IC50 and 2 IC50, respectively. The percentage of cells arrested in the Gap2/mitosis (G2/M) phase of the cell cycle increased from 8.48% (control) to 21.11% when the concentration of the extracts increased from 50 μg/ml to 100 µg/ml (Figure 4). When compared to the positive control (colchicine) (24.34%), it is extremely significant. In comparison to the positive control, colchicine (14.56%), approximately 9.54 % of them are currently arrested in the DNA synthesis (S) phase at this drug concentration (100µg/ml). For the positive control, colchicine, Sub-G0 was determined to be (0.36%), meaning that (0.36%) of the cell population was confirmed to be in apoptosis. The cells were gated at (0.2%) by the sample at 100 µg/ml. This has a significant impact (p<0.05).
Figure 4.
Histogram Displaying the Percentage of Gated Cells in Each Cell Cycle Phase as a Population. Image illustrating population histogram of the cells after treatment with extract. C, control (untreated); P, Positive control (25 µg/ml of Colchicine); E50, extract (50 µg/ml); E100, extract (100µg/ml). ** P<0.01, *** P<0.001.
Apoptosis Detection
As compared to the positive control (Colchicine) (43.11%), the viability percent decreased from 94.3% (control) to 54.35% at 100µg/ml, a highly significant difference (p<0.05), as shown in (Figure 5A and 5B). The amount of early apoptotic cells was found to have increased from 1.12% (control) to 21.34% (100µg/ml). Compared to the positive control (28.5%), a significant percentage of cells (22.05%) were driven into late apoptosis at 100µg/ml from 1.92% (control).
Figure 5.
A.Image depicting the fluorescence scatter diagram of the JURKAT line Annexin-V-FITC and PI stained cells are represented by the FSC-H and FL-2 heights, respectively. C: control (untreated); P: Positive control; E50: extract (50µg/ml); E100: extract (100µg/ml). B. Graph illustrating the flow cytometry analysis of apoptosis assay in JURKAT cells with and without treatment of extract. Colchicine (25µg/ml) was used as standard. * P<0.05, ** P<0.01, *** P<0.001.n.s. non-significant.
When JURKAT cells were treated to 50µg/ml of extract, they showed early apoptosis and a slight increase in late apoptosis. There were a few necrotic cells and a higher incidence of late apoptosis at a dosage of 100µg/ml. On the other hand, JURKAT cells showed increased levels of both early and late apoptosis when exposed to standard vincristine at a concentration of 25µg/ml. Orange apoptotic bodies indicates late apoptosis [30]. On the control slide, round green cells with a circular nucleus dispersed throughout the cell are visible (Figure 6). Red cells: Necrotic cells are those that have completely absorbed EtBr. Late apoptotic phase is depicted by yellowish-orange cells. Early-stage apoptotic cells are represented by green-yellow cells (Figure 6). The percentage of necrotic cells increased from 8 to 22% with increasing concentration to 100 from 50µg/ml. Positive control showed 34% necrotic cells. The percent of apoptotic cells as visible with the orange colour, was found to be 42, 10, and 46% respectively for positive, E50, and E100 (Figure 7).
Figure 6.
AO/EB Dual Staining Image Depicting the Quantification of Cell Cytotoxicity of Cells. C, control (untreated); P, Positive control; E50, extract (50µg/ml); E100, extract (100µg/ml).
Figure 7.
Graph Illustrating the Relative Fold Expression of Apoptotic Genes Caspase 3 and Caspase 8 in JURKAT Cell Line Post Treatment with Extracts at 50 and 100µg/ml. A, caspase 3; B, caspase 8. ** P<0.01, *** P<0.001.
Gene Regulation of Caspase 3 and Caspase 8
Compared to the control (1 or 100%), the relative expression of caspase 3 was found to be increased to 4.15 times at 100µg/ml. There was an uptick to 5.13 (p<0.05) in the positive control. Compared to the control (1 or 100%), the relative expression of caspase 8 was observed to be decreased 0.19 times at 100µg/ml. The drop to 0.09 (p<0.05) was observed in the positive control. Caspases 8 showed downregulation, while caspase 3 showed overexpression. According to the study, extracts alter how apoptotic genes are regulated in JURKAT cells. The caspase 3 gene was shown to be dose-dependently upregulated, while caspase 8 was found to be downregulated.
Discussion
Because endophytes may produce bioactive secondary metabolites which have potential use as biocontrol, antibacterial, anticancer and antioxidants, they are becoming more and more significant in the pharmaceutical and industrial domains [31]. In recent times, a great deal of research has been done on endophytes, mostly in temperate, humid continental climates that is, in tropical rainforests in Europe, America, and East Asia. The variety and content of endophytic microbiomes are determined by the ecological characteristics of the soil and host plant, such as the host plant’s interactions, environmental factors, and geographic location. To our knowledge, no previous reports for the presence of fungal endophytes in P. pinnata tree samples exist. Thus, the current study was conducted to separate endophytic fungi from P. pinnata and evaluate their beneficial secondary metabolites. We focused on the bark of P. pinnata trees because prior research has shown that it is extremely important for endophytes [32].
From the SBD plates, we could find two distinct types of fungi, of these, we discovered that Trichoderma species was more common and appeared more frequently (0.81) than Colletotrichum species (0.19). From the Pongamia species, numerous endophytic fungal species particularly Ascomycota and Trichoderma species were isolated. They act as opportunistic plant symbionts for disease management and yield enhancement, colonizing root surfaces and penetrating the first cell layers [33]. Trichoderma are endophytic plant symbionts that stimulate plant development, reduce the detrimental effects of plant disease, and enhance pepper and tomato seed germination [34]. According to Kashyap et al. [35], Trichoderma was also mostly observed in the rhizospheres of Pongamia species, which are antagonistic towards soil-borne diseases. We found our extracts from endophytes exhibited potent anticancer activity against JURKAT cell lines. We evaluated the cytotoxicity by MTT assay, acridine orange, flow cytometry and gene expression analysis. The study found that the extract and positive control significantly inhibited cell viability at concentrations of 21.26µg/ml and 48.65 µg/ml, respectively, compared to the control. The study found that the extract reduced cell viability significantly (IC50 48.65 µg/ml) compared to the control cells, indicating its anti-proliferating activity.
Numerous other distinct cytotoxic compounds derived from fungal endophytes have been discovered. To ascertain their precise mode of action against cancer, however, more cellular and molecular study is required [36]. These special molecules are chosen over conventional chemical treatments due to their high biodiversity, high absorption, safe effect on different tissues, and biocompatibility. They generally reflect the biological composition of lactones, alkaloids, quinones, and flavonoids. These secondary metabolites are thought to be a more dependable therapeutic alternative for cancer therapy because they demonstrated their effectiveness against several cancer cell lines [37]. Ever since the first discovery of Taxol (also called paclitaxel), a diterpenoid, from the endophytic fungus Taxomyces andreanae found in the bark of the Pacific Yew tree (Taxus brevifolia), it has been recognized that endophytic fungi are a valuable source of anticancer medications [38]. Subsequently, anticancer agents have been extracted from endophytic fungus, which include camptothecin from Entrophospora infrequens and 9-methoxycamptothecin and 10-hydroxycamptothecin from Fusarium solani. Additionally, the anticancer lead compounds podophyllotoxin from Phialocephala fortinii and deoxypodophyllotoxin from Aspergillus fumigatus [38] stimulated additional research on endophytic fungi, leading to the discovery of numerous other significant known and novel anticancer compounds. In the recent years, over 100 distinct fungus species have been found to date, producing over 200 possible anticancer compounds.
The MTT assay and LDH assay validated our results, showing a dose-dependent increase in LDH leakage with our extract, affecting JURKAT lines at 25 and 1000 µg/ml. We also found the concentration of extracts increased the percentage of cells arrested in the Gap2/mitosis phase from 8.48% to 21.11%, when compared to the positive (colchicine). At 100 µg/ml, approximately 9.54 % of cells were arrested in the DNA synthesis phase, indicating apoptosis. From our apoptosis assay, we found early apoptotic cells increased from 1.12% to 21.34% at 100µg/ml, while late apoptosis was driven into 22.05% of cells compared to the positive control. JURKAT cells showed early and late apoptosis when treated with extracts at 50µg/ml, with increased late apoptosis at 100 µg/ml, and increased early and late apoptosis at 25µg/ml. The study found that extracts alter the regulation of apoptotic genes in JURKAT cells, with caspase 3 gene being upregulated and caspase 8 downregulated. The relative expression of caspase 3 increased to 4.15 times at 100µg/ml, while caspase 8 decreased to 0.19 times. Similar results were reported about the potent anticancer agent of P. pinnata but from seeds [39, 40]. Many works were reviewed of the potential role of P. pinnata’s stem, leaves, seeds as anticancerous against various cancers which was well presented by using cell line models [41]. Even studies were reported of the potential role of trichoderma secondary metabolites as anticancerous, but no report were seen from P. pinnata. To our knowledge, ours would be the first report of the potential role of secondary metabolites extracted from endophytic fungus against JURKAT cell lines.
In conclusion, there hasn’t been a thorough investigation into the anticancer potential of endophytes from higher plant species, with particular attention to the P. pinnata plant. Studying them should be mandated immediately because the extinction of a plant specimen will also lead to the extinction of all related potential endophytes. Opportunities may arise from the global collection, cataloguing, and use of endophytic microbes. A rich and dependable source of natural compounds with intriguing biological activities, endophytic fungi also have a high level of biodiversity and can produce several compounds of pharmaceutical significance. These compounds are currently the focus of international scientific investigation as they are isolated and their potential for biotechnology is explored. In this work, we assessed the anticancer potential of secondary metabolites that were isolated from P. pinnata’s endophytic fungus species (Trichoderma). Assays for apoptosis, flow cytometry, MTT, and LDH all revealed positive antiproliferative activity. We have demonstrated experimentally that these extracts exhibited conformational stability and had the potential to function as anti-cancer drugs. Researchers can use the extracts’ in-vitro results as a powerful anticancer agent against JURKAT cell lines to guide in-vivo and in-vitro experimental studies for potential future uses.
Author Contribution Statement
Study conception and design by [Bassam Shaker Mahmood] and [Anwer Jaber Faisal], data collection by [Allaa Hatim Thanoon ] and [Baraa Ahmed Saeed], analysis and interpretation of results: [Ghassan M. Sulaiman] and [Sudhakar Malla.] All authors reviewed the report and approved the final version of the manuscript.
Acknowledgements
The authors thank Mustansiriyah University and the University of Technology, Baghdad, Iraq, for supporting this work.
If it was approved by any scientific Body/ if it is part of an approved student thesis
The manuscript was approved by the scientific committee of the Iraqi Center for Cancer and Medical Genetic Research, Mustansiriyah University, and is not part of any student thesis.
Ethics Statement
The study received ethical approval from the scientific committee of the Iraqi Center for Cancer and Medical Genetic Research, Mustansiriyah University, Baghdad, Iraq. Additionally, the study was conducted under the principles of the Declaration of Helsinki
Availability of data and materials
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author.
Conflict of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Verekar SA, Mishra PD, Sreekumar ES, Deshmukh SK, Fiebig HH, Kelter G, et al. Anticancer activity of new depsipeptide compound isolated from an endophytic fungus. J Antibiot. 2014;67:697–701. doi: 10.1038/ja.2014.58. [DOI] [PubMed] [Google Scholar]
- 2.Ding G, Song YC, Chen JR, Xu C, Ge HM, Wang XT, et al. Chaetoglobosin U, a cytochalasan alkaloid from endophytic Chaetomium globosum IFB-E019. J Nat Prod. 2006;69(2):302–4. doi: 10.1021/np050515+. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Fu W, Zhu LZ, Liu XF, Li L, Peng LZ, et al. Anti-tumor effects and mechanism of a novel camptothecin derivative YCJ100. Life Sci. 2022;311:p121105. doi: 10.1016/j.lfs.2022.121105. [DOI] [PubMed] [Google Scholar]
- 4.Sonowal S, Gogoi U, Buragohain K, Nath R. Endophytic fungi as a potential source of anti-cancer drug. Arch Microbiol. 2024;206(3):p122. doi: 10.1007/s00203-024-03829-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Zhang QY, Jia M, Ming QL, Yue W, Rahman K, et al. Endophytic fungi with antitumor activities: their occurrence and anticancer compounds. Crit Rev Microbiol. 2014;42:454–73. doi: 10.3109/1040841X.2014.959892. [DOI] [PubMed] [Google Scholar]
- 6.Saeed BA, Faisal AJ, Mahmood BS, Thanoon AH. Chronic inflammation induced by Escherichia coli blood infections as a risk factor for pancreatic cancer progression. Asian Pac J Cancer Prev. 2024;25(12):4407–14. doi: 10.31557/APJCP.2024.25.12.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S, Sharma B, Kanwar S, Kumar A. Lead phytochemicals for anticancer drug development. Front Plant Sci. 2016;7:1667. doi: 10.3389/fpls.2016.01667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–77. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 9.Kusari S, Hertweck C, Spiteller M. Chemical ecology of endophytic fungi: origins of secondary metabolites. Chem Biol. 2012;19:792–8. doi: 10.1016/j.chembiol.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Demain AL, Vaishnav P. Natural products for cancer chemotherapy. Microb Biotechnol. 2011;4:687–99. doi: 10.1111/j.1751-7915.2010.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph B, Priya RM. Bioactive compounds from endophytes and their potential in pharmaceutical effect: a review. Am J Biochem Mol Biol. 2011;1:291–309. [Google Scholar]
- 12.Lei M, Ribeiro H, Kolodin G, Gill J, Wang YS, Maloney D, et al. Establishing a high-throughput and automated cancer cell proliferation panel for oncology lead optimization. J Biomol Screen. 2013;18:1043–53. doi: 10.1177/1087057113491825. [DOI] [PubMed] [Google Scholar]
- 13.Salvador-Reyes LA, Luesch H. Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat Prod Rep. 2015;32:478–503. doi: 10.1039/c4np00104d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stierle A, Strobel G, Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;260:214–6. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- 15.Balunas MJ, Kinghorn AD. Drug discovery from medicinal plants. Life Sci. 2005;78:431–41. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Karamala LN, Yalpi K, Megha R, Aditi N, Rachana V, Prasanna A, et al. Exploring the therapeutic potential of Pongamia pinnata plant extract against skin cancer: in-silico and in-vitro study. J Ethnopharmacol. 2025;337:118964. doi: 10.1016/j.jep.2024.118964. [DOI] [PubMed] [Google Scholar]
- 17.Anuradha R, Krishnamoorthy P. Screening of phytochemicals and identification of chemical constituents of Pongamia pinnata by GC-MS. Chem Technol. 2012;4:16–20. [Google Scholar]
- 18.Wu Z, Ameer K, Jiang G. Isolation and characterization of anti-tumor compounds from ethyl acetate extract of Rumex japonicus Houtt roots and their cytotoxic effects. Food Sci Technol. 2021;42:e05421. [Google Scholar]
- 19.Tibpromma S, Karunarathna S, Bhat J, Suwannarach N, Stephenson S, Elgorban A, et al. Using culture-dependent and molecular techniques to identify endophytic fungi associated with tea leaves (Camellia spp ) in Yunnan Province, China. Diversity. 2022;14:287. [Google Scholar]
- 20.Rajabi M, Moghadam M, Azizi A, Soltani J. Isolation and molecular identification of two rutin-producing endophytic fungi from Caper (Capparis spinosa L ) Biol J Microorg. 2022;11:169–80. [Google Scholar]
- 21.Ali-Rachedi F, Meraghni S, Touaibia N, Mesbah S. Analyse quantitative des composés phénoliques d’une endémique algérienne Scabiosa atropurpurea sub maritima L. Bull Soc R Sci Liege. 2018;87:13–21. [Google Scholar]
- 22.Kim D-O, Chun OK, Kim YJ, Moon H-Y, Lee CY. Quantification of polyphenolics and their antioxidant capacity in fresh plums. J Agric Food Chem. 2003;51:6509–15. doi: 10.1021/jf0343074. [DOI] [PubMed] [Google Scholar]
- 23.Wei AH, Montesinos P, Ivanov V, DiNardo CD, Novak J, Laribi K, et al. Venetoclax plus LDAC for newly diagnosed AML ineligible for intensive chemotherapy: a phase 3 randomized placebo-controlled trial. Blood. 2020;135:2137–45. doi: 10.1182/blood.2020004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia Y, Xu T, Wang C, Li Y, Lin Z, Zhao M, Zhu B. Novel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapy. Int J Nanomedicine. 2018;13:143–59. doi: 10.2147/IJN.S148960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jhaumeer Laulloo S, Bhowon MG, Soyfoo S, Chua LS. Nutritional and biological evaluation of leaves of Mangifera indica from Mauritius. J Chem. 2018;10:6869294. [Google Scholar]
- 26.Ciniglia C, Pinto G, Sansone C, Pollio A. Acridine orange/ethidium bromide double staining test: a simple in-vitro assay to detect apoptosis induced by phenolic compounds in plant cells. Allelopathy J. 2010;26(2):301–8. [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Mahmood BS. Silver nanoparticles and their role in gene expression of motility gene motB and repression of AI-2-controlled gene. Baghdad Sci J. 2020;17(3):916. [Google Scholar]
- 29.Faisal AJ, Kadhim MJ, Abbood AS, Jarallah ET. Molecular detection of genital pathogens in semen from infertile and fertile men. Acta Microbiol Bulg. 2025;41(1):57–62. [Google Scholar]
- 30.Nakkala JR, Mata R, Sadras SR. Green synthesized nano silver: synthesis, physicochemical profiling, antibacterial, anticancer activities and biological in vivo toxicity. J Colloid Interface Sci. 2017;499:33–45. doi: 10.1016/j.jcis.2017.03.090. [DOI] [PubMed] [Google Scholar]
- 31.Elsayed HE, Kamel RA, Ibrahim RR, Abdel-Razek AS, Shaaban MA, Frese M, et al. Cytotoxicity, antimicrobial, and in silico studies of secondary metabolites from Aspergillus sp isolated from Tecoma stans (L ) Juss ex Kunth leaves. Front Chem. 2021;9:760083. doi: 10.3389/fchem.2021.760083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akram M, Nimesh S, Chishti MA, Ahmad MI, Dhama S, Lal M. Pongamia pinnata: an updated review on its phytochemistry and pharmacological uses. Pharm Pharmacol Int J. 2021;9(5):194–9. [Google Scholar]
- 33.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2(1):43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 34.Marlina A, Hasanah N. Efforts to control Fusarium oxysporum wilt disease using biological agents FMA fungus and Trichoderma harzianum. Floratek. 2011;6(8):8–17. [Google Scholar]
- 35.Kashyap PL, Rai P, Srivastava AK, Kumar S. Trichoderma for climate resilient agriculture. World J Microbiol Biotechnol. 2017;33(8):1–18. doi: 10.1007/s11274-017-2319-1. [DOI] [PubMed] [Google Scholar]
- 36.Adeleke BS, Babalola OO. J Fungi (Basel) Pharmacological potential of fungal endophytes asociados with medicinal plants: a review;7:147. doi: 10.3390/jof7020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amirzakariya BZ, Shakeri A. Phytochemistry. Bioactive terpenoids derived from plant endophytic fungi: an updated review (2011-2020);197:113130. doi: 10.1016/j.phytochem.2022.113130. [DOI] [PubMed] [Google Scholar]
- 38.Beheshti F, Shabani AA, Akbari Eidgahi MR, Kookhaei P, Vazirian M, Safavi M. Evid Based Complement Alternat Med. Anticancer activity of Ipomoea purpurea leaves extracts in monolayer and three-dimensional cell culture;2021:6666567. doi: 10.1155/2021/6666567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh VP, Yedukondalu N, Sharma V, Kushwaha M, Sharma R, Chaubey A, et al. Lipovelutibols A–D: cytotoxic lipopeptaibols from the himalayan cold habitat fungus Trichoderma velutinum. J Nat Prod. 2018;81(2):219–26. doi: 10.1021/acs.jnatprod.6b00873. [DOI] [PubMed] [Google Scholar]
- 40.Jabir MS, Yosif HM, Hasoon BA, Rasool KH, Jawad KH, Ali IA, et al. Au@ hydroxyapatite nanocomposite as a novel apoptosis inducer and NF-κB translocation inhibitor in prostate cancer cell line. Inorg Chem Commun. 2024;170:113469. [Google Scholar]
- 41.Abbas JA, Hasoon BH, Jabir MS. Insights into the apoptosis induction in colon cancer cells by Ag-Cu bimetallic nanocomposite: 2D tissue culture model and insilico study. Inorg Chem Commun. 2025:114019 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author.