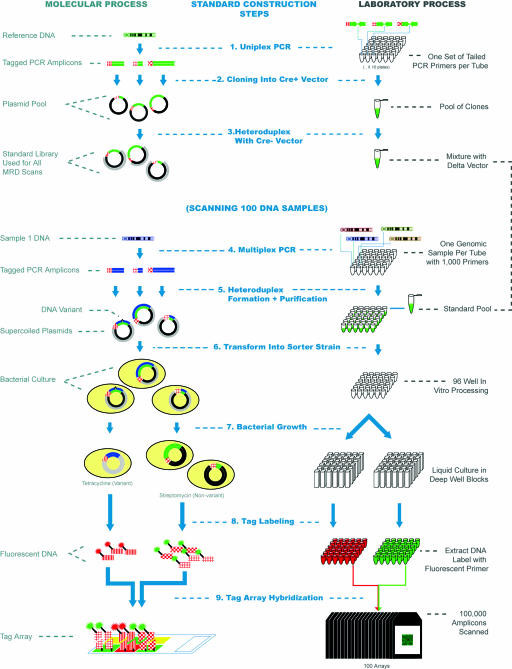
Fig. 1.
Schematic of the MRD on tag arrays process. A molecular representation of the various steps in the process is shown on the left, whereas the corresponding laboratory process is shown on the right (1). The process starts with the generation of a set of plasmid standards for each of the sequences to be scanned. PCR amplicons are generated for each target by using tailed primers that allow tag sequences to be incorporated. Colors are used to distinguish DNA from different genomic sources (DNA from the reference genome is green). The red textured line segments represent different tag sequences, whereas individual PCR amplicons are shown as being of different lengths, to distinguish them (2). The PCR amplicons are cloned into the cre+ vector (shown as black), pooled, and mixed (3) with digested cre- vector (shown in gray) to create a pool of standards in a single tube that are used for all individual MRD scans (4). PCR amplicons are then generated from each DNA sample under investigation by using the same priming sequences as for the standards. In this process, >1,000 amplicons are generated for each unique DNA sample in each well of a 96-well plate (5). The PCR products are denatured and then mixed with the standard pool. Heteroduplex molecules are then formed between the unknown DNA sequences and the standard DNA. The nicks are closed by using a ligase enzyme, and the supercoiled plasmids are purified in a 96-well format. Each well then contains the heteroduplex plasmids from 1,000 amplicons for a single individual (6). Material from each well is then used to transform the mutation sorter strain (7). The resulting culture is split into two deep 96-well plates. Streptomycin is added to one and tetracycline to the second, and the bacteria are grown as liquid cultures overnight to complete the separation of variant and nonvariant plasmids (8). Plasmid DNA is then extracted from the cultures in 96-well format, and run-off reactions with a labeled primer are used to add a fluorescent label. The primers used to amplify DNA from the tetracycline cultures have a 6-carboxyfluorescein (FAM) group (arbitrarily shown as red), and those used to amplify the streptomycin cultures have a biotin group that ultimately binds to streptavidin-phycoerythrin (9). Finally, the two labeled DNAs from each sample are pooled and hybridized to the universal tag microarray. A total of 96 arrays are used to scan the 96 DNA samples under investigation. The fluorescence intensities of each tag feature allow the variation status of each of the 1,000 amplicons to be determined. Steps (4)-(9) are then repeated for each set of 96 DNA samples in the experiment.