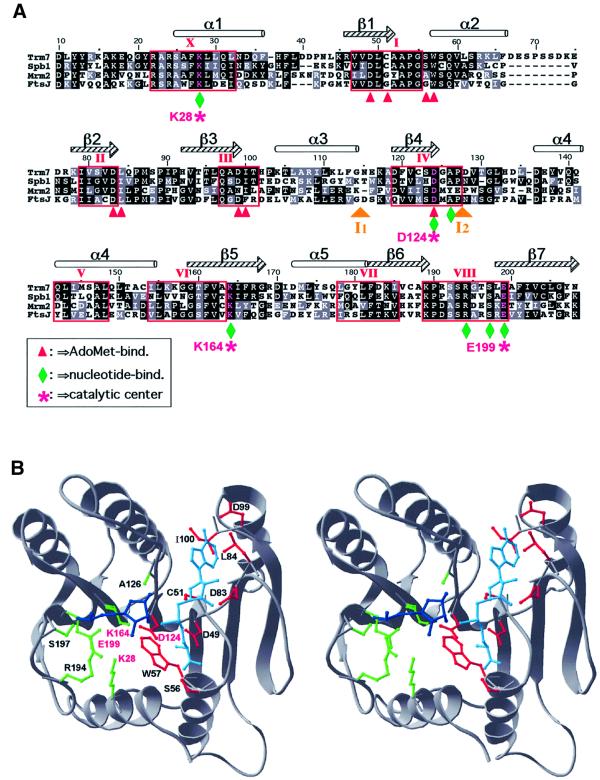
Fig. 2. Three-dimensional structure prediction for Trm7p. (A) Alignment of the AdoMet-binding domain of the three yeast proteins (Trm7p, Spb1p and Mrm2p) with the E.coli protein FtsJ/RrmJ. Identical and chemically equivalent residues are highlighted in black and grey, respectively. Secondary structural elements are indicated above the alignment (α-helices as cylinders and β-strands as arrows). Conserved motifs have been labelled according to the nomenclature proposed by Posfai et al. (1988). Predicted interactions with AdoMet and the methylated nucleotide are designated by red arrowheads and green diamonds, respectively. The predicted catalytic tetrad K–D–K–E is labelled with purple stars. Two insertions in Mrm2p [I1 (58 amino acids) and I2 (21 amino acids)] have been omitted for the sake of clarity. (B) The stereogram of the Trm7p model in cartoon representation. The AdoMet moiety is shown in cyan, and the ribose and the phosphate group of the nucleotide to be methylated are shown in blue. The predicted binding and catalytic residues are shown in the wireframe representation, with colour coding analogous to that in (A).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.