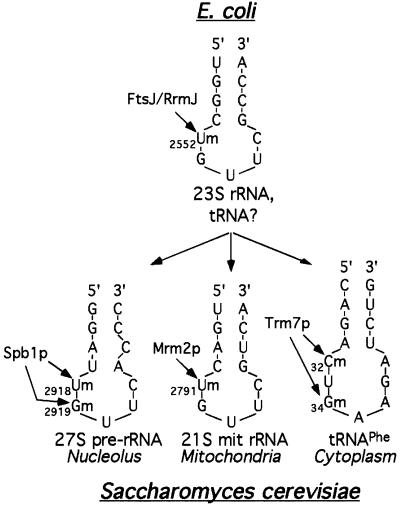
Fig. 9. Schematic representation of the secondary structure of the target sites for each of the three enzymes that belong to the 2′-O-RNA MTase family. Only the 2′-O-ribose methylations discussed in this report are represented. Top: location of U2552 in the hairpin/loop of domain V of the peptidyl transferase centre of the E.coli 23S rRNA. In addition, FtsJ/RrmJ can also methylate tRNA in vitro; however, the position of the methyl-nucleotides has not yet been determined (Bügl et al., 2000). Bottom, left: location of Um2918 and Gm2919 in the hairpin/loop of the peptidyl transferase centre of the nuclear 27S pre-rRNA. Bottom, middle: location of Um2791 in the hairpin/loop of the 21S mitochondrial rRNA. Bottom, right: location of C32 and G34 in the anticodon loop of tRNAPhe. The modified nucleotides are indicated by arrows, with their known (FtsJ/RrmJ, Trm7p, Mrm2p) or putative (Spb1p) modification enzymes.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.