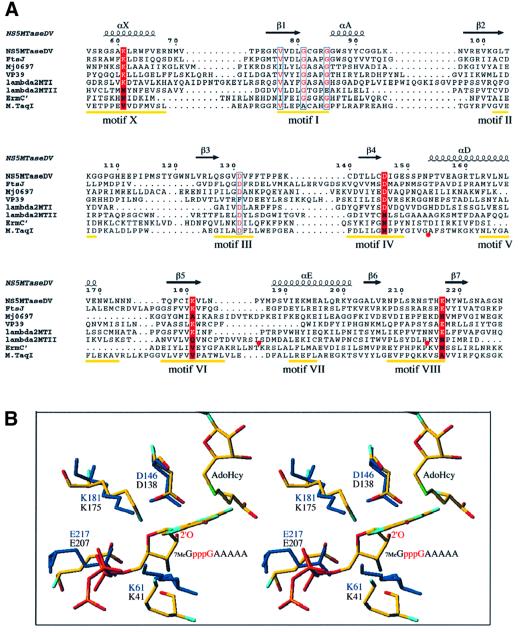
Fig. 5. Comparative analysis of NS5MTaseDV. (A) Structure-based sequence alignment of the MTase core domain of NS5MTaseDV with rRNA MTase FtsJ (Bugl et al., 2000), the C-terminal rRNA MTase domain of Mj0697 (Wang et al., 2000), mRNA MTase VP39 (Hodel et al., 1996), mRNA MTase domains I and II of Reovirus protein λ2 (Reinisch et al., 2000), the rRNA MTase ErmC′ (Bussiere et al., 1998) and the DNA MTase M.TaqI (Schluckebier et al., 1997). The positions of conserved DNA MTase motifs I to X [except for motif IX, which is only conserved for (cytosine-5-) DNA MTases; Posfai et al., 1989] are indicated in yellow (Malone et al., 1995). In order to simplify the alignment, additional structural elements in the sequences of λ2 MTase II (between residues Ile936 and Ser961 as well as Arg989 and Lys1014) and in M.TaqI (between residues Gly112 and Ala132) were omitted. The omissions are indicated by red circles below the sequences. The alignment was done manually using Seaview (Galtier et al., 1996) according to the superposition of the structures using TURBO (Roussel and Cambillau, 1991), and the final form of the alignment was generated with ESPript (Gouet et al., 1999). The secondary structure elements of the NS5MTaseDV core are given above the alignment and are named as in Figure 1B. The residues within DNA MTase motifs I and III, presenting similarity as detected by ESPript, are shown in red with blue outlines. Positions conserved between NS5MTaseDV, FtsJ, VP39 and λ2 MTase I, which coincide with motifs X, IV, VI and VIII, are marked by a red background where the conserved active-site residues K-D-K-E of 2′OMTases are in white. (B) Stereo view of the superposition of active site residues K-D-K-E of VP39 and NS5MTaseDV. Both structures are shown as sticks. The VP39 structure (PDB accession code 1AV6) is depicted with a RNA substrate (7MeG-capped RNA hexamer) and AdoHcy. VP39 residues K41-D138-K175-E207, AdoHcy and the RNA substrate are coloured according to atom type (oxygen in red, nitrogen in blue, carbon in yellow, sulfur in green and phosphorous in orange; hydrogens are not shown). The target 2′-OH group of the second nucleotide of the substrate is annotated (only a pppG fragment of the substrate 7MeGpppGAAAAA is depicted). NS5MTaseDV residues K61-D146-K181-E217 are shown in dark blue, AdoHcy was omitted.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.