Abstract
We have studied developmentally regulated patterns of histone acetylation at high resolution across ∼54 kb of DNA containing three independently regulated but neighboring genetic loci. These include a folate receptor gene, a 16 kb condensed chromatin region, the chicken β-globin domain and an adjacent olfactory receptor gene. Within these regions the relative levels of acetylation appear to fall into three classes. The condensed chromatin region maintains the lowest acetylation at every developmental stage. Genes that are inactive show similarly low levels, but activation results in a dramatic increase in acetylation. The highest levels of acetylation are seen at regulatory sites upstream of the genes. These patterns imply the action of more than one class of acetylation. Notably, there is a very strong constitutive focus of hyperacetylation at the 5′ insulator element separating the globin locus from the folate receptor region, which suggests that this insulator element may harbor a high concentration of histone acetylases.
Keywords: acetylation/boundaries/chromatin domains/folate receptor/globin genes
Introduction
In the eukaryotic nucleus transcribed regions are organized into independently regulated domains, each containing the necessary regulatory elements for the correct expression of the genes located within (Bell et al., 1998; Bell and Felsenfeld, 1999). There is abundant evidence that chromatin, in addition to playing a structural role in the organization of DNA within the nucleus, is also intimately involved in the regulation of eukaryotic gene expression (Felsenfeld, 1996; Wu, 1997; Kornberg and Lorch, 1999; Farkas et al., 2000; Schubeler et al., 2000). Associated with this regulatory function are a variety of histone modifications, including acetylation, phosphorylation, methylation, ADP ribosylation, ubiquitylation and glycosylation, that can alter the structure and chemical reactivity of chromatin (Cheung et al., 2000; Strahl and Allis, 2000).
A great deal of attention has been focused on histone acetylation. There have been numerous studies of the mechanisms of acetylation and deacetylation, and their consequences for gene expression (Grunstein, 1997; Turner, 2000). These studies have established that, particularly in the neighborhood of transcriptionally active promoters and enhancers, lysine residues on the N-terminal tails of histones H3 and H4 are more highly acetylated than those in regions of transcriptional inactivity (Kuo et al., 1998). Many transcription factors bound at promoters and enhancers can recruit histone acetylases, explaining how acetylation levels can be changed in the neighborhood of such sites (Bannister and Kouzarides, 1996; Mizzen et al., 1996; Ogryzko et al., 1996; Chen et al., 1997). Recent work has also suggested how acetylation signals might be propagated over the chromatin of coding regions, for example by the attachment of certain acetylases to the moving transcription complex (Wittschieben et al., 1999).
The N-terminal tails of histones H3 and H4 both have several lysines residues capable of being acetylated. The fact that different acetylases and deacetylases target specific lysines suggests that acetylation of specific lysines may have important regulatory functions (Strahl and Allis, 2000). Among studied systems and organisms, the modification of individual lysine residues shows considerable variation. In Drosophila and Saccharomyces cerevisiae, Lys12 of H4 appears to be specifically acetylated in heterochromatin (Turner et al., 1992; Braunstein et al., 1996), whereas all H4 lysines are unacetylated in mammalian facultative heterochromatin (Jeppesen and Turner, 1993). In studies of a CMVlacZ transgene incorporated into HeLa cells, acetylation of lysines 5, 12 and 16 of H4 was shown to be involved in the initiation of chromatin opening, whereas acetylation of Lys8 appeared to be important for the maintenance of open chromatin (Chen and Townes, 2000). In S.cerevisiae an increase in acetylation of H4 Lys5 appears to be sufficient for derepression of UME6-regulated genes in rpd3 deletion mutants (Rundlett et al., 1998). Although such studies show that individual lysines can have distinct regulatory roles, it is not clear whether these are the same from one gene or organism to another.
Even the correlation between transcriptional activity and overall acetylation levels of H3 and H4 differs between the systems that have been investigated. Some studies show nearly identical levels of H3 and H4 acetylation enrichment over transcriptionally active genes (Parekh and Maniatis, 1999; Elefant et al., 2000). In contrast, in a recent study of the human β-globin locus, H4 acetylation was uniformly elevated over the locus, whereas the greatest enrichment in H3 acetylation occurred over two observed hypersensitive sites and the β-globin gene (Schubeler et al., 2000).
Our laboratory has been interested in the relationship between chromatin structure and gene expression in the neighborhood of the chicken β-globin locus. The β-globin locus itself is >33 kb long, and includes four globin genes that are developmentally regulated and expressed in erythroid cells (Felsenfeld, 1993) (Figure 1). A region of ‘condensed’ chromatin extends ∼16 kb upstream of the globin locus. Beyond that we have identified another gene, coding for a folate receptor (FR), which is expressed only in immature erythroid cells, before the globin genes are active (Prioleau et al., 1999). An important feature of this region is the presence of a boundary element, or insulator, at the 5′ end of the globin locus, just 3′ of the beginning of the condensed chromatin domain (Chung et al., 1993). It has been shown (Hebbes et al., 1994) that in 15-day-old embryonic erythrocytes the transition between the nuclease-sensitive, highly acetylated chromatin of the globin domain and the insensitive, underacetylated chromatin of the condensed region occurs just 5′ of the insulator.
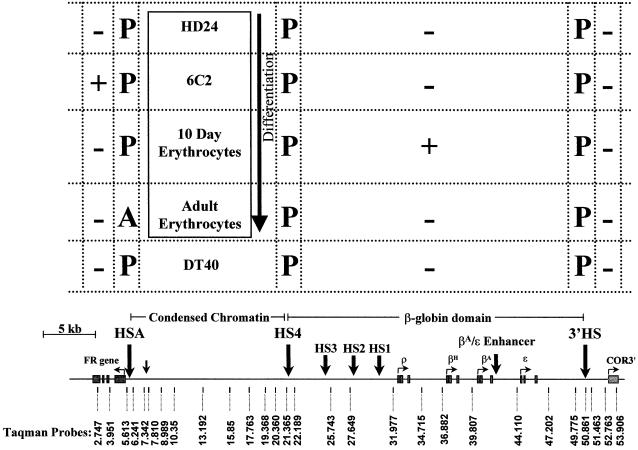
Fig. 1. Expression and chromatin structural changes in a 54 kb region containing the FR gene, β-globin gene and chicken olfactory receptor gene in cell lines derived from different erythropoietic stages and a non-erythroid cell. In the cells or cell lines listed, + or – indicates whether the gene in the map below is expressed or silent, respectively. P or A indicates the presence or absence of DNase I-hypersensitive sites. A scale map of the FR gene, condensed chromatin region, β-globin domain and chicken olfactory receptor gene is shown at the bottom. The locations of hypersensitive sites (HSA, HS4 and 3′HS) are indicated by large arrows. ‘Taqman probes’ denotes the name and location of primer pairs and Taqman probes used for analysis.
There are major differences in chromatin structure, DNA methylation state and gene expression patterns within the 54 kb encompassing the FR–globin gene regions in cells arrested at various stages of erythroid development (Clark et al., 1993; Felsenfeld, 1993; Prioleau et al., 1999). At the earliest erythroid stage examined neither FR nor any of the globin genes is expressed. At a slightly later stage only FR is expressed, and still later FR is silent and globin gene expression is turned on. We wanted to understand how these changes were correlated with alterations in chromatin structure, and particularly with histone acetylation states. We therefore undertook a quantitative study, at very high resolution, of the pattern of histone acetylation across the entire region in cells corresponding to each of these developmental stages. We show that patterns of acetylation are very different from one stage to another, but are consistent with the variation in patterns of gene expression. The results indicate that both site-specific and domain-wide mechanisms of acetylation must be at work, and that the boundary element between the FR and the globin genes is a site of permanent hyperacetylation. This suggests possible mechanisms of boundary action.
Results
Acetylation patterns of cells with different programs of expression
In earlier work (Prioleau et al., 1999) we made use of chicken embryonic erythroid cells and well characterized chicken cell lines, arrested at two pre-differentiation developmental stages, to correlate chromatin structural changes and DNA methylation patterns with gene expression. In HD24 lines, both the FR gene and all of the globin genes are silent. In 6C2 cells, corresponding to a slightly later developmental stage, the FR gene is expressed but globin genes are not. In 10 day circulating erythrocytes, the adult globin genes are expressed but FR expression is shut off (Figure 1).
We wished to measure the acetylation levels of the core histones H3 and H4 in each kind of cell, at closely spaced sites across the entire region. For this purpose we made use of antibodies raised against histone tails acetylated at either single or multiple sites. As in previous studies by others (Hebbes et al., 1994; Elefant et al., 2000), micrococcal nuclease (MNase) digests of nuclei from each cell type were purified on sucrose gradients, and the monomer and dimer fractions were pooled for use in immunoprecipitation experiments. These contained predominantly monomers. This assured that the resolution of the assay would be at the level of mono- or dinucleosomes (see Materials and methods).
Previous experiments have analyzed the enrichment of the immunoprecipitated (IP) fractions by slot blot or quantitative PCR (Hebbes et al., 1994; Elefant et al., 2000; Schubeler et al., 2000). We chose to analyze this 54 kb region using Taqman real-time PCR. The speed of the method and the ability to measure a large number of samples simultaneously allowed us to employ probes targeted at relatively small intervals across the domain, and to study multiple cell lines. In this method, the exonuclease activity of Taq polymerase used in the PCR attacks a specifically annealed Taqman probe, resulting in liberation of a fluorescent reporter (Figure 2A). The number of PCR cycles necessary to reach a predetermined fluorescence intensity is a measure of the initial abundance of the target sequence. The probes chosen were for regions free of sequence repeats, and an attempt was made to space them evenly across the 54 kb region (Figure 1, map). Additional probes were added to examine DNase I-hypersensitive sites, the FR gene, β-globin genes and chicken olfactory receptor 3′ β1 gene (COR3′) (Figure 1, map).
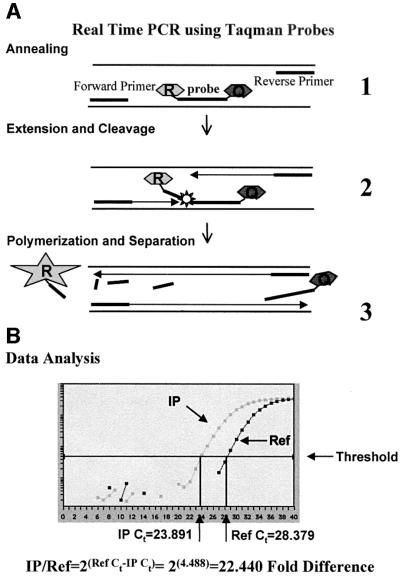
Fig. 2. Real-time PCR using Taqman probes. (A) A schematic of PCR in the presence of a Taqman probe. (1) Probes and primers anneal to target sequence. Taqman probes have two covalently linked fluorescent dyes: a reporter (R) and a quencher (Q). On the probe, the reporter dye emission is quenched. (2) During each extension cycle, the 5′→3′ exonuclease activity of Taq DNA polymerase cleaves the reporter dye from the probe. (3) Once separated from the quencher, the reporter dye emits its characteristic fluorescence, which is measured in every cycle by the ABI Prism 7700 sequence detector. (B) A representative panel of data generated by the ABI Prism 7700 sequence detector software for Taqman probe 5.613 on samples immunoprecipitated from 6C2 nuclei. IP indicates the signal for the fluorescent reporter detected in each cycle for the immunopreciptated target sequence. Ref indicates a similar analysis of the target sequence abundance in total input DNA. Ct indicates the cycle threshold number. Below the data panel is a sample calculation to determine the fold difference between the IP sample and the reference sample.
In order to determine the enrichment or depletion of the immunoprecipitation for a specific antibody, the total amount of DNA in the IP fraction was measured, and an equal amount of input DNA (before immunoprecipitation) was run in parallel as a reference standard. The relative abundance of each sequence was determined from the difference in the number of PCR cycles necessary to reach a fixed signal threshold value. Enrichment of a sequence in the IP fraction was calculated from the difference of the threshold cycle number (Ct) for the IP fraction to the Ct for the input DNA. A schematic presentation of the analysis is shown in Figure 2B.
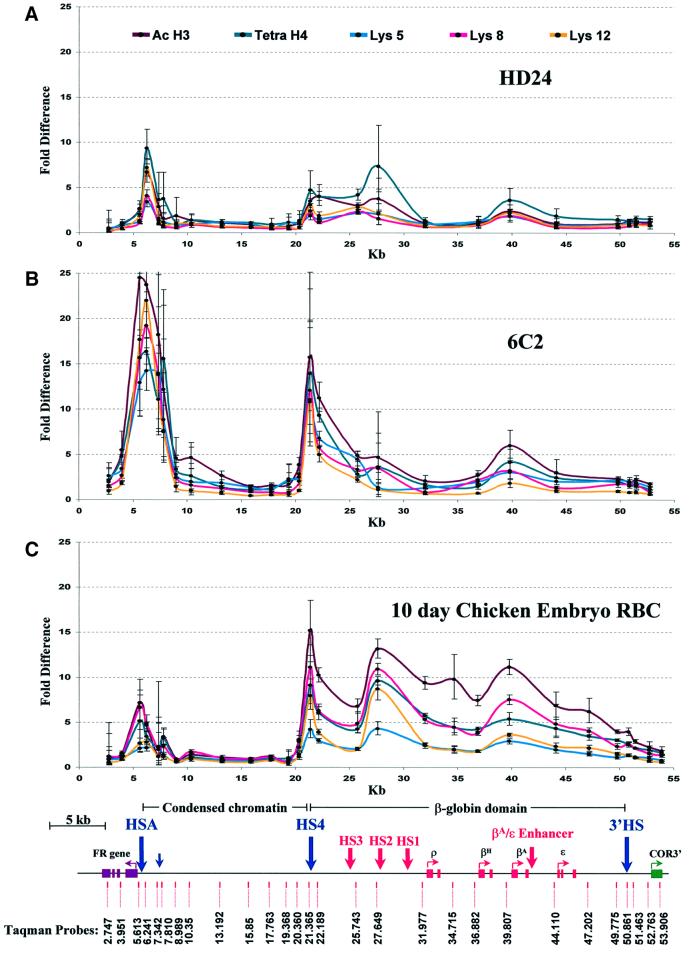
Data from the analysis of erythroid cells at three different stages are given in Figure 3. Each data point represents the ratio of abundance of that probed sequence in the IP fraction to its abundance in the input chromatin fraction before immunoprecipitation. There are dramatic changes in the patterns of acetylation accompanying progression from early HD24 cells to the more mature erythrocyte stage. In HD24 cells (Figure 3A), there are only low levels of acetylation throughout much of the region. The notable exceptions are: (i) peaks of acetylation in the neighborhood of 5′HS4, the insulator element that marks the 5′ boundary of the β-globin locus, and 3′ of HSA, just upstream of the FR promoter region; and (ii) small peaks over 5′HS2 in the β-globin locus control region (LCR) and near the adult β-globin promoter. The 16 kb condensed chromatin region is quite unacetylated. Major changes in these patterns are seen in 6C2 cells, which are arrested at a later stage when FR is expressed. There is now a very large peak of acetylation over HSA, but also extending toward the coding region itself, for H3 and lysines 5, 8 and 12 of H4. The effect is to move the peak of acetylation upstream (i.e. 5′ on the map) of HSA, directly over the FR promoter. However, probes within the body of the FR gene, the first intron (3.951) and last exon (2.747), show low levels of acetylation (Figure 3B). There is also strong acetylation at all histone sites measured over the insulator region (5′HS4), as well as significant enrichment over the promoter of the βA-globin gene. In both these and HD24 cells, an enhancer blocking element (3′HS), which marks the 3′ boundary of the β-globin locus and the COR3′ gene, has little or no observable acetylation. The 16 kb condensed chromatin region is once again largely unaffected by the hyperacetylation of the surrounding regions.
Fig. 3. Acetylation across three independently regulated domains of two erythroid lines and 10 day embryonic erythroid cells at different stages of development. The graphs show the relative chromatin immunoprecipitation of acetylated histone H3, acetylated histone H4 and specific acetylated histone H4 lysines of mono and di-nucleosomes from HD24 cells (A), 6C2 cells (B) and 10 day chicken embryo red blood cells (C). The y-axis indicates the fold difference between the input fraction and the bound fraction and the x-axis indicates position in kb relative to the end of the FR gene. Different line colors (top) correspond to acetylation of individual sites (H4 lysine 5, 8 or 12) or multi-acetylated H3 or H4. The lower section shows the name and location of primer pairs and Taqman probes used for analysis. Several DNase I-hypersensitive sites are indicated: HSA, HS4, HS3, HS2, HS1 and 3′HS.
The pattern of acetylation in 10 day chicken embryo red blood cells (Figure 3C) is different from that in the two cell lines. FR gene expression is no longer observed in these cells, and the level of acetylation over that gene has now returned essentially to baseline, though some acetylation (comparable to that seen in HD24 cells) is seen over HSA. In contrast, acetylation levels over 5′HS4 are quite high, and this extends over the entire β-globin domain, in confirmation of the earlier observations (Hebbes et al., 1994). This widespread increase in acetylation over the globin genes contrasts with the narrow region of acetylation observed in 6C2 cells over the FR promoter. Although these high levels can be seen at all histone acetylation sites observed, they appear in the case of the β-globin region to be greatest for H3. Once again the 16 kb condensed chromatin region is, within the limits of our assay, totally unacetylated.
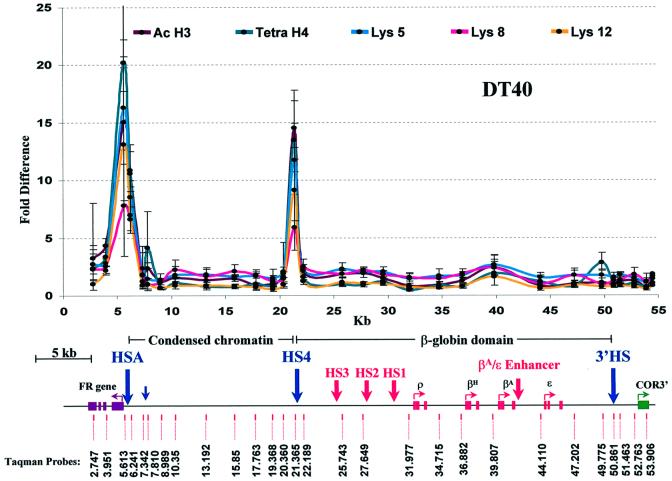
We compared the erythroid lineage patterns with those observed in a chicken lymphocyte cell line (DT40) (Figure 4). In these cells, neither FR nor any of the globins is expressed (Prioleau et al., 1999). There is no significant acetylation over either the globin locus or the FR gene. Nevertheless, very strong peaks of H3 and H4 acetylation are observed both at HSA and 5′HS4, which flank the unacetylated 16 kb condensed chromatin region.
Fig. 4. Acetylation in a non-erythroid cell line. The graph shows the relative chromatin immunoprecipitation of acetylated histone H3, acetylated histone H4 and specific acetylated histone H4 lysines of mono- and dinucleosomes from DT40, a chicken lymphocyte cell line. Designations and map are the same as in Figure 3.
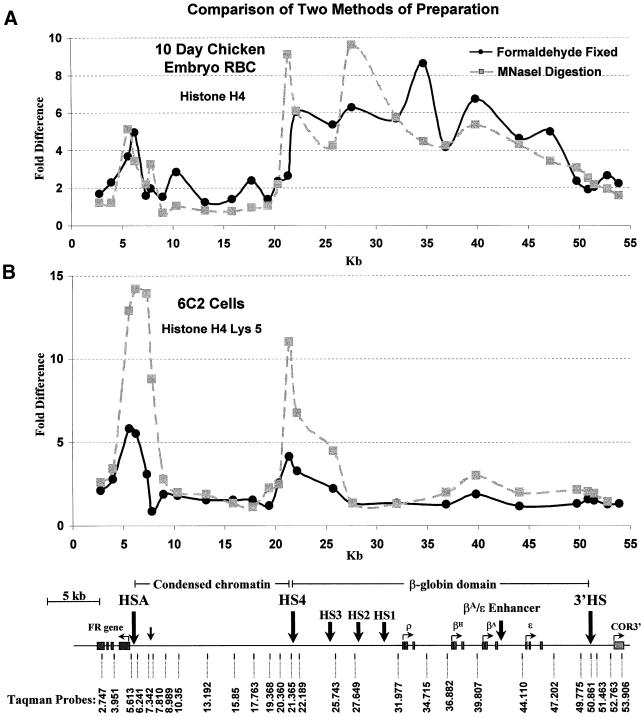
The results in Figure 3 were obtained with a fractionated pool of mono- and dinucleosomes generated by MNase digestion (see Materials and methods). We followed procedures for nuclear isolation that avoided formaldehyde fixation, similar to methods described previously (Hebbes et al., 1994) for measurement of histone acetylation levels. We compared our results with those obtained by a different procedure (Orlando et al., 1997), involving direct formaldehyde fixation of cells and sonication of chromatin (see Materials and methods). This also avoids the use of a deacetylation inhibitor during isolation of nuclei. Formaldehyde treatment was adjusted carefully to give high yields of cross-linked material that would also yield relatively small molecular weight sonication products. It did not prove possible to obtain a preponderance of mono- or dinucleosomes by this procedure. Nonetheless, the results of immunoprecipitation experiments with these fractions isolated from 10 day chicken embryo erythrocytes show features similar to those seen with our standard procedure. This similarity is clearly seen when the results for formaldehyde-fixed chromatin from 10 day chicken red blood cells is compared directly with the data from Figure 3 for chromatin prepared from the same cells by MNase digestion (Figure 5A). Although differences are observed, the general trend of elevated levels of acetylation over the globin domain and adjacent to the FR promoter is maintained between the two methods. The principal difference between the results obtained with the two methods lies in the reduced prominence of the peaks 3′ of HSA and 5′ of HS4 in the cross-linked material. In particular the peak over HS4 appears to be shifted downstream. This is the result expected when the precipitated fragments are larger, so that results are averaged over several nucleosomes. Resolution is thus impaired by cross-linking, and in fact, the shifted behavior can be generated from the MNase digestion results by averaging the data over approximately five nucleosomes, the estimated size of the fragments generated in the cross-linked sample. We also carried out immunoprecipitations with cross-linked chromatin from 6C2 cells (Figure 5B). These results are again similar to those obtained by MNase digestion (Figure 3B), with the expected broadening and lowering of the peaks seen with cross-linked material. Here, however, as in the case of DT40 cells (Figure 4), the globin genes are not being transcribed. As a result the peak over HS4 is clearly visible against the surrounding negative background. Histones within the HS4 boundary element continue to be acetylated, consistent with the conclusion that HS4 is a constitutively acetylated site.
Fig. 5. Two methods of preparation for chromatin used in immunoprecipitation of acetylated histones show similar patterns. (A) Comparison of the relative acetylation of histone H4 measured following two methods of preparation in 10 day chicken embryo erythrocytes. (B) The relative acetylation of Lys5 on histone H4 in 6C2 cells, compared for two different methods of chromatin preparation. In both (A) and (B), the black line shows data for chromatin that was fixed with 1% formaldehyde and sonicated to generate small fragments. The gray dashed line (data from Figure 3) shows results for chromatin prepared from nuclei digested with MNase and purified on a sucrose gradient to isolate mono- and dinucleosomes. Designations and map are the same as in Figure 3.
The fraction of histones acetylated
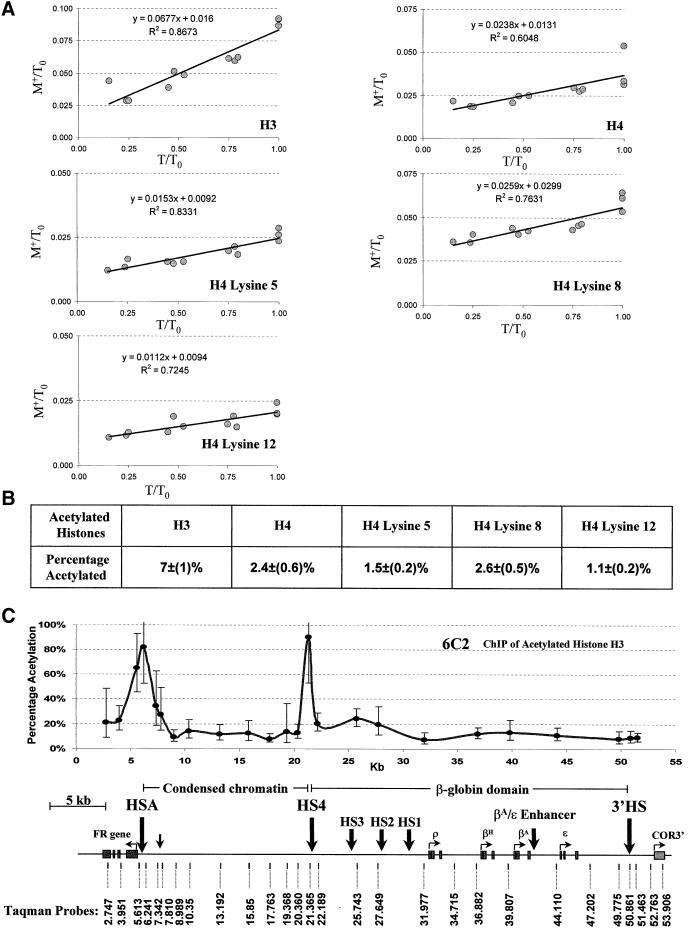
The availability of high-resolution data makes it reasonable to attempt some quantitative analysis. The immunoprecipitations described above were carried out in chromatin excess over antibody. Each data point in Figure 3 reflects enrichment of a particular sequence in the acetylated fraction relative to its abundance in the input sample. An additional number, the fraction of input chromatin precipitable in antibody excess, is needed to calculate for each point within the FR–globin–COR3 locus the fraction of nucleosomes acetylated at a particular lysine residue (see Materials and methods for details). We devised conditions to carry out this measurement without using large quantities of antibody. A fixed amount of a given antibody was titrated with decreasing amounts of chromatin, with antibody always in excess. The total DNA content before precipitation and the total DNA content of the precipitate were measured for each data point using a picogreen assay and plotted as a fraction of total chromatin (the largest amount of total DNA content measured before precipitation; see Materials and methods). Regression analysis of the data (Figure 6A) gave the fraction of all material that contained lysines acetylated at the positions recognized by the antibody.
Fig. 6. Percentage of acetylated nucleosomes in total chromatin immunoprecipitated with specific antibodies. (A) Determination of the maximum percentage of immunoprecipitated nucleosomes acetylated at specific histones or histone sites. Each tube contains a fixed amount of antibody, always in excess over chromatin. Increasing amounts of chromatin are added and the fraction of IP material, M+, is measured (see Materials and methods). T is the amount of input chromatin at each point in the titration. T0 is the amount of chromatin in the tube with the highest concentration. The immunoprecipitate (M+) as a fraction of T0 (M+/T0) is plotted against the input chromatin (T) as a fraction of T0 (T/T0). The line represents an average of data from separate titrations of the same antibody. The percentage of acetylated nucleosomes in total chromatin for a specific histone or histone sites is determined from the value of the slope (see Materials and methods). The linearity and non-zero slope of the plots are consistent with the presence in each tube of an antibody excess. This does not preclude the possibility that a sub-population of acetylated nucleosomes is not precipitable by the antibody. The fact that the line does not go through the origin shows that there is a fixed amount of material that precipitates in each tube. (B) A table of the percentage of acetylated nucleosomes in total chromatin. Specific histones and histone H4 acetylation sites are indicated above. The maximum percentage of nucleosomes immunoprecipitated by a specific anti-acetyl-histone antibody is indicated immediately below. (C) The absolute percentage of acetylation of histone H3 in 6C2 cells across the 54 kb region. The y-axis indicates the percentage of acetylation. All other designations are the same as in Figure 3.
We have carried out this analysis for 6C2 cells with all of the antibodies (Figure 6B). The antibody against acetylated histone H3 (AcH3) precipitated the largest fraction of material (∼7%). We carried out on this same chromatin sample a separate analysis of the enrichment in AcH3 across the entire FR–globin–COR3 locus. Using the number for the total precipitable material, we can now approximate (see Materials and methods) the absolute abundance of acetylated nucleosomes throughout the locus (Figure 6C). At the largest peak, which lies over HSA, nearly every nucleosome (82%) is acetylated. Obviously a single recognized acetylation event on a nucleosome is sufficient for it to be immunoprecipitated, so this does not allow us to conclude that every site is fully acetylated, although this is certainly consistent with the results.
Discussion
In this study, we examined the levels of acetylation across a ∼54 kb region of the chicken genome, containing three independently regulated systems of genes, a region of condensed chromatin devoid of genes, at least two boundary elements and multiple regulatory elements. We took advantage of the availability of two well characterized cell lines, HD24 and 6C2, which are arrested at different stages of erythroid development, and have long been used as model systems for the study of these stages. We also used erythroid cells taken from 10-day-old chick embryos, and a non-erythroid cell line, DT40. This allowed us to survey the state of acetylation of a variety of chromatin structures in a series of environments, and particularly to follow the changes that are correlated with altering patterns of expression over the region. Our results show that there are separate domains of acetylation, each with its own distinct program.
In HD24 cells, which represent early stage erythroid cells (BFU-E), none of the genes—FR, β-globin or COR3′—is expressed. Nonetheless there are centers of acetylation over regulatory elements and hypersensitive sites. The chromatin over HSA, near the FR, is most highly acetylated, with strongest peaks corresponding to H3, H4 and H4 K12. Acetylation is also seen over the insulator element (HS4), the globin LCR element 5′HS2 and the adult β-globin promoter. The region of condensed chromatin that separates the peaks at HSA and 5′HS4 is not acetylated at any detectable level in our assay. It should be noted that HSA and 5′HS4 are DNase I-hypersensitive sites in all of the cell lines we describe here, except for adult erythrocytes. (Figure 1).
The FR gene is actively transcribed at a specific stage in early erythroid development (Prioleau et al., 1999). Among the cell lines or populations we have studied here, FR expression is confined to 6C2 cells (CFU-E). The acetylation pattern in these cells reflects this activity. There is very strong acetylation over HSA, which contains important regulatory elements for the FR (M.Simpson, unpublished). Interestingly, this does not extend over the body of the FR gene, which is not significantly acetylated here or at any other stage. Similar high acetylation levels are seen over 5′HS4. The pattern over the rest of the globin locus, and in the condensed chromatin region, is similar to that seen in HD24. In the case of 6C2, we have additional quantitative information (Figure 6C) allowing us to conclude that almost all of the nucleosomes over HSA and the preponderance of those over 5′HS4 are acetylated, while other peaks correspond to only partial acetylation.
A strikingly different pattern is observed in 10 day embryonic erythrocytes. Here, consistent with earlier observations, we find very high levels of acetylation distributed rather uniformly over the domain carrying the β-globin genes. Many of the probes used for this region include promoters and enhancers, but at this developmental stage the genes encoding βH-globin and βA-globin are expressed and the other genes are not active (Felsenfeld, 1993). Furthermore, two probes (at 34.715 and 47.202) are targeted to sequences well separated from any known enhancer or promoter, and these also detect high acetylation levels. We conclude that at least some of the acetylases responsible are not operating from tethered positions centered on promoters and enhancers, but are capable of functioning over considerable distances within the region. It is now well established that some acetylases can be part of an RNA polymerase transcription complex that allows processive acetylation (Wittschieben et al., 1999). In addition, it has been reported that there are two classes of acetylase, one behaving as tethered enzymes able to act only on neighboring nucleosomes, the other able to acetylate over considerable distances (Vignali et al., 2000).
It is also instructive to compare these results with the data for the bursal lymphocyte cell line DT40 (Baba et al., 1985). In these cells there is no expression of FR, globins or COR3′ (Bulger et al., 1999; Prioleau et al., 1999). Correspondingly, there is no significant acetylation over any of these genes, nor over any of the upstream elements of the globin LCR (HS1–HS3). Nonetheless, there is strong acetylation over HSA and 5′HS4. These sites thus appear to be constant centers of acetylation in all of the cells we have studied. They flank a region of condensed chromatin that is unacetylated with equal constancy.
Examination of the patterns of acetylation for H3, H4 and the individual H4 lysine residues does not reveal any consistent and striking differences between them. Unlike results for the human β-globin locus (Schubeler et al., 2000), there is no clear distinction between the patterns of histones H3 and H4. In both cases there is enrichment over HSA and 5′HS4, and a parallel increase in acetylation over the globin locus in cells where the globin genes are either active or about to be activated developmentally. Multiple measurements were made at each site (see Materials and methods), and although in some cases the error bars indicate that differences may occur, these differences are generally small. We are therefore reluctant to ascribe to them any physiological significance.
The insulating properties of the DNA sequences surrounding 5′HS4 have been well studied (Chung et al., 1993; Bell et al., 1999). A 1.2 kb sequence containing the hypersensitive site can act as a positional enhancer blocker, preventing an enhancer from activating a promoter when it is placed between them. When surrounded by two copies of the 1.2 kb fragment, a stably integrated reporter is protected against position effects (Chung et al., 1993; Pikaart et al., 1998). There is also evidence that a 250 bp ‘core’ element is able to confer the same protection (F.Recillas-Targa, unpublished). Within the core, the enhancer blocking activity has been correlated with a binding site for the regulatory factor CTCF (Bell et al., 1999), but other sequences in the core are required for protection against position effects (F.Recillas-Targa, unpublished). It has been suggested that the globin 5′HS4 element serves to insulate the FR and the globin genes from each other, ensuring that the strong regulatory elements for FR at HSA and for the globin genes at HS1–HS3 do not cross-activate the inappropriate gene during development (Prioleau et al., 1999). In addition, the 5′ insulator at HS4 may prevent encroachment of the upstream condensed chromatin region onto the β-globin domain.
For these reasons it is quite interesting that the 5′HS4 region is a center of histone acetylation in all cells that we have examined. We proposed earlier (Pikaart et al., 1998) that this insulator might confer protection against position effects by serving as a source of histone acetylases, which maintained acetylation levels within a protected domain. The strong peak of acetylation over 5′HS4 does indicate that it is a focus of acetylation activity, consistent with such a model. We note that the same is true for the chromatin near HSA, in the FR promoter, and we suggest that another border element may be present there. In contrast to the strong centers of acetylation at 5′HS4 and HSA, the CTCF binding site at 3′HS (Saitoh et al., 2000) just upstream of the COR3′ gene, is not markedly modified, although small peaks can be seen in 10 day embryonic erythrocytes and DT40 cells. This suggests that CTCF binding alone is not sufficient to attract acetylases, and focuses our attention on the proteins bound at adjacent sites in the 5′ boundary.
Our results show that a region ∼54 kb long may display within it a wide variety of acetylation patterns, and that these may change with the state of activation of the individual genes it contains. We observe three classes of behavior. The condensed chromatin region remains completely unacetylated in every cell type we have examined. This is certainly consistent with the known properties of this region, but it is interesting that there is a tendency for the ends of this region to acquire high levels of acetylation. A second class of behavior is exhibited by the globin and FR genes. As expected, the acetylation of these regions is closely correlated with levels of expression. The positive regulatory elements, notably some of the upstream locus control elements of the β-globin LCR, show signs of acetylation in HD24 cells that precede the activation of expression seen in 10 day embryonic erythrocytes. The hypersensitive sites at opposite ends of the condensed chromatin region display the third kind of behavior. As noted above, these are hyperacetylated in all of the lines we have studied. It will be interesting to determine whether these sites play a role in establishing or maintaining the programmed variation in acetylation patterns that we have observed.
Materials and methods
Cells
HD24 and 6C2 cells were maintained as described in Prioleau et al. (1999). DT40 cells are an avian leukosis virus-induced bursal lymphoma cell line derived from Hyline SC chicken (Baba et al., 1985). These cell lines were purchased from American Type Culture Collection (ATCC, Rockville, MD) and grown in DMEM supplemented with 50 mM β-mercaptoethanol, 2 mM glutamine, 10% fetal bovine serum, 5% chicken serum and 10% tryptose phosphate broth, with antibiotics. Ten day chicken erythrocytes were obtained by bleeding 10 day embryos from fertilized White Leghorn chicken eggs (Truslow Farms, Chestertown, MD).
Micrococcal digestions and preparation of di- and mononucleosomes
Nuclei for HD24, 6C2 and DT40 cells, and 10 day chicken embryo erythrocytes were prepared as described (Prioleau et al., 1999). All buffers were supplemented with 10 mM Na-butyrate, 0.4 mM phenylmethylsulfonyl fluoride (PMSF) and 10 µg/ml aprotinin. Di- and mononucleosomes were prepared as described (Hebbes et al., 1994). Except where obtaining representative fractions of chromatin from highly condensed and highly open regions, three equal aliquots of nuclei were digested with increasing concentrations of MNase. The MNase activity was adjusted so that the lowest concentration generated chromatin fragments larger than trinucleosomes, the middle concentration generated mainly tri-, di- and mononucleosomes, and the highest concentration generated principally mononucleosomes. All MNase digestions were incubated for 10 min at 37°C and stopped by adding EDTA pH 8.0 to a final concentration of 10 mM. The three digests were combined and di- and mononucleosomes were purified on a 5–30% sucrose gradient (Hebbes et al., 1994).
Formaldehyde cross-linking and sonication
Chromatin fixation, purification and sonication were carried out as described (Orlando et al., 1997). Briefly, 1 × 108 cells were fixed for 1 min at 25°C with 1% formaldehyde in 25 ml of culture media such that sonication (Misonix Ultrasonic processor XL) with a 2 mm tip at maximum setting for 180 s (10 s on, 10 s off) generated DNA fragments from ∼200 to 2000 bp. Approximately 1 × 107 cells were used for chromatin immunoprecipitations, which were carried out as described below except that washes, elution and reversal of cross-linking were identical to those described (Orlando et al., 1997).
Chromatin immunoprecipitation
Chromatin immunoprecipitations were carried out as described (Hebbes et al., 1994). Briefly, to reduce non-specific binding to protein A, 700 µl of chromatin, prepared as described above, were incubated with 100 µl of protein A–agarose (50% slurry in ChIP buffer) for 1 h at 4°C with gentle rocking. ChIP buffer is 50 mM NaCl, 10 mM Tris–HCl pH 7.4, 10 mM Na butyrate, 1 mM EDTA, 0.1 mM PMSF, 10 µg/ml aprotinin. Millipore ultrafree-MC 0.45 µm Microfuge filter units were used to separate protein A–agarose and chromatin. At this point DNA was prepared from a sample of protein A-purified chromatin and used as the ‘input’ sample. Antibodies specific for acetylated histones (100 µl) [anti-acetylated histone H3, anti-acetylated histone H4, anti-acetyl-histone H4 (Lys5), anti-acetyl-histone H4 (Lys8) and anti-acetyl-histone H4 (Lys12) supplied by Upstate Biotechnology] were incubated with protein A-clarified chromatin for 2 h at 4°C while rocking gently. One hundred microliters of protein A–agarose were added to the chromatin antibody mix and incubated overnight at 4°C while rocking gently. The protein A– agarose and unbound chromatin were separated using 0.45 µm Microfuge filter units. The first filtrate was kept as the unbound sample. The protein A–agarose was washed five times with ChIP buffer and the bound chromatin was eluted, first with 250 µl of ChIP buffer plus 1.5% SDS, and then with 250 µl of ChIP buffer plus 0.5% SDS. DNA from bound and unbound chromatin was purified by phenol/chloroform extraction and ethanol precipitation. DNA samples were quantified using picogreen (Molecular Probes, Eugene, OR) fluorescence.
Determination of the fraction of nucleosomes acetylated in the genome
To determine the maximum precipitable chromatin for a given antibody, 10, 5, 2.5 and 1.25 µg of micrococcal mononucleosomes were immuno precipitated with 50 µl of the appropriate anti-acetyl-histone antibody, an excess in each case, as described above. Each chromatin immunoprecipitation was repeated three times. Total DNA and IP DNA were quantified using picogreen fluorescence. The measured DNA immunoprecipitated in each sample, M+, can be described as M+ = Tg + B, where T is the total amount of DNA in each sample measured before immunoprecipitation, and g is the true fraction of chromatin precipitable. B is a measure of the constant background of precipitated material that we observe. We plot M+/T0 versus T/T0 where T0 is the amount of chromatin in the tube with the highest chromatin concentration (Figure 6A). This gives a line with slope equal to g, the constant fraction of chromatin that can be precipitated under conditions of antibody excess (Figure 6A).
Real-time PCR and data analysis
Differences in DNA concentration obtained from the immunoprecipitation of acetylated and unacetylated histone fractions were determined by real-time PCR using the ABI Prism 7700 sequence detector following PE Applied Biosystem’s Taqman Universal PCR Master Mix protocol. Real-time PCR was carried out in triplicate on 2 ng of DNA at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Data were collected at 60°C. The concentration of primers and Taqman probes used was determined by following the optimization procedure described in PE Applied Biosystem’s protocol. For each experiment, the threshold was set to cross a point at which real-time PCR amplification was linear (Figure 2B). For the majority of the experiments, data were analyzed with a threshold of 0.05. Data collected was analyzed and plotted using Microsoft Excel. The fold difference of a given target sequence precipitated by a specific antibody was determined by dividing the amount of target sequence in the IP fraction by the amount of target sequence in input DNA (Ref). Briefly, for any PCR, amplification can be described by the formula X = Xo(1 + E)n where Xo is the initial DNA concentration of a target sequence, X is the final DNA concentration of a target sequence, E is the efficiency (a number from 0 to 1) and n is the number of cycles. Therefore, if a threshold is set at which a specific final DNA concentration X is reached, X is a constant, and the number of cycles (Ct) required to reach X is inversely related to the initial target sequence concentration Xo, i.e. Xo(IP)/Xo(Ref) = [X(IP)/X(Ref)](1 + E)Ct(IP)–Ct(Ref). If X(Ref) = X(IP), which should be true for the same primer and Taqman probe set, and E = 1, then Xo(IP)/Xo(Ref) = 2Ct(IP)–Ct(Ref). Therefore, for each Taqman probe, the ratio of IP to Ref is calculated by subtracting the Ct determined for the target sequence of the IP sample from the Ct determined for the target sequence of the reference sample and taking the resulting power of 2 (Figure 2B).
Primers and Taqman probes
Primers and Taqman probes were selected from the chicken β-globin domain, 16 kb condensed chromatin and FR gene sequence using PE Applied Biosystems Primer Express software. Primer and Taqman probes were obtained from Gibco-BRL and PE Applied Biosystems, respectively. For each Taqman probe, PCR products were designed to be <147 bp (the size of a nucleosome monomer) with an average size of 86 bp. In a few cases (probes 2.747, 6.241, 7.810, 10.350 and 21.365), the choice of effective priming sequences required separations of 121–144 bp. In these cases, therefore, much of the signal will be contributed by the di- and trinucleosomes in the sample population. The primers and Taqman probes used are given in Table I.
Table I. Taqman probes and primers used.
| Taqman probe | Primers |
|---|---|
| 2.747 | 5′-CCTGGCAGGAGAGGATCTCTT |
| 5′-CCAACTCCTACAAATACACCACAGAA | |
| 5′-6FAM-TCAAACCACATCTGGATCAGGCGC-TAMRA | |
| 3.951 | 5′-CAGCAAAGAGTCCTTTCCCTTCT |
| 5′-TTCAACACCATCTTGCTGCAAT | |
| 5′-6FAM-CAAAACACCCGAGGGCTGCCTG-TAMRA | |
| 5.613 | 5′-AACATTACCTGCCTAGAGACTATCCA |
| 5′-CTGTGTCAGAAGGCTTTCCTGTTA | |
| 5′-6FAM-CCACAACAACACTCAGAACAGCAGCCTC-TAMRA | |
| 6.241 | 5′-GGGTCCGACCAGGAAGGA |
| 5′-TCAGTGCCAGGATTGAAGCA | |
| 5′-6FAM-ACAGACCAGCAGATCTTCCTATTGGCACA-TAMRA | |
| 7.342 | 5′-CACAGCACTGCAGCAGCATT |
| 5′-ATCCAAAAACAAACCCGATATCA | |
| 5′-6FAM-CTCTGTTTTCTGTTCCTCGCCGAGGA-TAMRA | |
| 7.810 | 5′-CCCAGCATACAGGACACCATT |
| 5′-CACAAAGACCAGTCCCTCCAAT | |
| 5′-6FAM-CGCCTGGGTTCCGATCCCATG-TAMRA | |
| 8.989 | 5′-GGGCCCAATGAACCAGAAA |
| 5′-TGTTCCCCAGCAACGCA | |
| 5′-6FAM-AAATGGCAAACTGTTGATGGGAACGC-TAMRA | |
| 10.350 | 5′-GGAACAAGTTGGCAAGGTCCTAT |
| 5′-TCTTCTGCCCTGCCCGTAT | |
| 5′-6FAM-TGCAGTTCCCTGTTCATGTGCTTTTCG-TAMRA | |
| 13.192 | 5′-GAATGTGTCCATCTGCCCTCAT |
| 5′-GGGAAGCCATCCCTGCA | |
| 5′-6FAM-TGCTGAGCATGTGGCTGCCTCC-TAMRA | |
| 15.850 | 5′-CAGCAGACGCTGTGGTGAA |
| 5′-CTTGCAGGATGCAGACTGGA | |
| 5′-6FAM-ATCCCATCGGTGCCACCCTGAG-TAMRA | |
| 17.763 | 5′-TGTTATCGCACACACACACACTTT |
| 5′-GACAGGGATGTTCTTCCTCTGAA | |
| 5′-6FAM-TCGTCTGATGCAGACAATTTCTCTGTGATCTC-TAMRA | |
| 19.368 | 5′-TGCTGGCAGTCACACGTCA |
| 5′-AACGAAAACACAATAAATAATAGCAATGA | |
| 5′-6FAM-TCCAGGCTTGGCCGAGCATCC-TAMRA | |
| 20.360 | 5′-GCTCTGCGAGGGCTCTCTTT |
| 5′-CCTCTCCTCACCCACCTGTTT | |
| 5′-6FAM-TTCCCGCTCTCTGTTAATATTGGATTTCCTTTTT-TAMRA | |
| 21.365 | 5′-CTCTGTGCTCAGCATCCTTCAAT |
| 5′-CCTTTCGGCACTTTCTTCCTTT | |
| 5′-6FAM-CTCCGCTGCACCTCCTCTGCAAA-TAMRA | |
| 22.189 | 5′-CAGGACAGCATGGACGTGG |
| 5′-TTCTGAACGCTGTGACTTGGA | |
| 5′-6FAM-CATGCAGGTGTTGAGGCTCTGGACA-TAMRA | |
| 25.743 | 5′-GCCCGTGCTGTTTGCAC |
| 5′-TGAGTCACGGTTGTGTGTGGT | |
| 5′-6FAM-AGCCGTGTTATCGCCCCATGGC-TAMRA | |
| 27.649 | 5′-GCCCCACCAGCCGTG |
| 5′-CATCACCGCTGCCCAAA | |
| 5′-6FAM-TGTGTTCACTTGGTGCCACTGTCAGC-TAMRA | |
| 27.729 | 5′-CCCCTGCACCACCTCTCTC |
| 5′-GGCATCTATGGGAAAACTGAGTTT | |
| 5′-6FAM-TCCCTGCGCAAAACGACGAGAGA-TAMRA | |
| 31.977 | 5′-CAGAGGAGCCAACATTTGGG |
| 5′-CCCCTCTGGGTGATGCATT | |
| 5′-6FAM-CGCTGCAGGCGTGAAGCCATT-TAMRA | |
| 34.715 | 5′-CGTGTTCTGGAGGAGAGAGAAGA |
| 5′-CTTATCAGCAAGACTGGCAGATGT | |
| 5′-6FAM-TGACTGGCTGTGGTCCAGAGGCTG-TAMRA | |
| 36.882 | 5′-TGACACTGGAAACCTATGGCC |
| 5′-AGCCCCGAGTGCAGGTG | |
| 5′-6FAM-TGGAGGAGCCATGCAGGCAGC-TAMRA | |
| 39.807 | 5′-CTGTGGTCTCCTGCCTCACA |
| 5′-AGGCTGGGTGCCCCTC | |
| 5′-6FAM-CAATGCAGAGTGCTGTGGTTTGGAACTG-TAMRA | |
| 42.112 | 5′-CCTTGTTTATGCACTTCTTCACCC |
| 5′-AACCCCCTCTCTTCCCTCAC | |
| 5′-6FAM-CGCTGCCCATTCTGCTGCTCTG-TAMRA | |
| 44.110 | 5′-TGGATGTGGTGCTCAGGGAT |
| 5′-CAACCACAACCCGACCAAAC | |
| 5′-6FAM-TGATTTAGCGGAGGGCTGTTAGAATTAGGGC-TAMRA | |
| 47.202 | 5′-GATGGGAAAACCCTGAACCTC |
| 5′-CAACCTGCTAGAGAAGATGAGAAGAG | |
| 5′-6FAM-CGCTGCACTGAATGGACGGGAA-TAMRA | |
| 49.775 | 5′-TTGGGATTCCCCTGCTGAT |
| 5′-GCCTACATTTTGGCTGTGCA | |
| 5′-6FAM-CAAAAAATGATCGCACATCACCACGTG-TAMRA | |
| 50.861 | 5′-TTCACAAAACACCAGTTATGCTCC |
| 5′-ACCTGCTGCTTCAGAGGCA | |
| 5′-6FAM-TCTGCTGGTGAGATGGCGTCTGCT-TAMRA | |
| 51.463 | 5′-GGCAGATGGAGTGTGAGTTCC |
| 5′-GGCTGCAGTAGCTGATGGG | |
| 5′-6FAM-TGAGTGCTGAGGGCTGGGAAATTTTG-TAMRA | |
| 52.763 | 5′-CCCTGACGGAGATGAGGATC |
| 5′-CCCTTCTCCTGGTCCCCAT | |
| 5′-6FAM-CTTGTCTGGGCAGGAGAGCAGTGTGA-TAMRA | |
| 53.906 | 5′-TCTGTGCCGTCCTCATCTTCT |
| 5′-GCATTCCTTCCATACCGGTG | |
| 5′-6FAM-CATCCCTTTGGCTGGGCTGTCCA-TAMRA |
Analysis of data
We want to estimate what fraction of sites at a target locus actually carry an acetyl group. With the PCR techniques used here, we measure the fold difference in sequence abundance between the IP fraction and the input sample, which is effectively the ratio R, where R = (moles of target sequence in IP acetylated fraction/moles of total IP DNA)/(moles of target sequence in input DNA/moles of total input DNA). If we define Ftarget = moles of target sequence in IP acetylated fraction/moles of target sequence in input DNA and Ftotal = moles of total IP DNA/moles of total input DNA, then R = Ftarget/Ftotal. In Figures 3 and 4, the values of the enrichment factor R are plotted as a function of target position in the locus.
If the target site is fully acetylated (Ftarget = 1) then R = 1/Ftotal. Therefore, the fraction of total DNA immunoprecipitated places an upper limit on the observed value of R. (Obviously, if all of the chromatin is fully acetylated, there can be no enrichment of any particular sequence in the IP fraction.) Running the argument in reverse, if the measured value of R is RM, then, in the general case where only part of the genome is acetylated, Ftotal must be ≤(1/RM). For example, if a 10-fold enrichment is measured for H4 K12 at the target sequence, then ≤10% of all genomic nucleosomes in the input can be acetylated at H4 K12. The ratio R does not tell us directly what percentage of all target nucleosomes in the genome is acetylated (the fractional site occupancy). The immunoprecipitation conditions bring down only a small fraction of all acetylated nucleosomes. To calculate the occupancy we must also know what percentage of all nucleosomes is acetylated. This requires an immunoprecipitation experiment in which an excess of antibody is used. The fractional occupancy can now be calculated for every other point on the graph in Figure 3; it will be proportional to the measured value of R at that point. For example, if ∼7% of genomic nucleosomes can be acetylated at H3 (as seen in Figure 6B), then the upper limit for the fold enrichment is RM = 1/Ftotal = 1/0.07 = 14.3. Consequently, the fraction of nucleosomes acetylated in Figure 6C at the site detected by probe 6.241 (where R = 11.7) is 11.7/14.3 or 0.82. Thus, ∼82% of nucleosomes at that site are acetylated.
Acknowledgments
Acknowledgements
We would like to thank Michael Pikaart and Colyn Crane-Robinson for their advice and assistance.
References
- Baba T.W., Giroir,B.P. and Humphries,E.H. (1985) Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology, 144, 139–151. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bell A.C. and Felsenfeld,G. (1999) Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev., 9, 191–198. [DOI] [PubMed] [Google Scholar]
- Bell A., Boyes,J., Chung,J., Pikaart,M., Prioleau,M.N., Recillas,F., Saitoh,N. and Felsenfeld,G. (1998) The establishment of active chromatin domains. Cold Spring Harb. Symp. Quant. Biol., 63, 509–514. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–396. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M. et al. (1999) Conservation of sequence and structure flanking the mouse and human β-globin loci: the β-globin genes are embedded within an array of odorant receptor genes [published erratum appears in Proc. Natl Acad. Sci. USA (1999) 96, 8307]. Proc. Natl Acad. Sci. USA, 96, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- Chen W.Y. and Townes,T.M. (2000) Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl Acad. Sci. USA, 97, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Tanner,K.G., Cheung,W.L., Sassone-Corsi,P., Denu,J.M. and Allis,C.D. (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Clark D., Reitman,M., Studitsky,V., Chung,J., Westphal,H., Lee,E. and Felsenfeld,G. (1993) Chromatin structure of transcriptionally active genes. Cold Spring Harb. Symp. Quant. Biol., 58, 1–6. [DOI] [PubMed] [Google Scholar]
- Elefant F., Cooke,N.E. and Liebhaber,S.A. (2000) Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem., 275, 13827–13834. [DOI] [PubMed] [Google Scholar]
- Farkas G., Leibovitch,B.A. and Elgin,S.C. (2000) Chromatin organization and transcriptional control of gene expression in Drosophila. Gene, 253, 117–136. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. (1993) Chromatin structure and the expression of globin-encoding genes. Gene, 135, 119–124. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. (1996) Chromatin unfolds. Cell, 86, 13–19. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. and Turner,B.M. (1993) The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell, 74, 281–289. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D. and Lorch,Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Zhou,J., Jambeck,P., Churchill,M.E. and Allis,C.D. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C.A. et al. (1996) The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Parekh B.S. and Maniatis,T. (1999) Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell, 3, 125–129. [DOI] [PubMed] [Google Scholar]
- Pikaart M.J., Recillas-Targa,F. and Felsenfeld,G. (1998) Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev., 12, 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau M.N., Nony,P., Simpson,M. and Felsenfeld,G. (1999) An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J., 18, 4035–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Bell,A.C., Recillas-Targa,F., West,A.G., Simpson,M., Pikaart,M. and Felsenfeld,G. (2000) Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J., 19, 2315–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Francastel,C., Cimbora,D.M., Reik,A., Martin,D.I. and Groudine,M. (2000) Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev., 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Turner B.M., Birley,A.J. and Lavender,J. (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell, 69, 375–384. [DOI] [PubMed] [Google Scholar]
- Vignali M., Steger,D.J., Neely,K.E. and Workman,J.L. (2000) Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J., 19, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben B.O. et al. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell, 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Wu C. (1997) Chromatin remodeling and the control of gene expression. J. Biol. Chem., 272, 28171–28174. [DOI] [PubMed] [Google Scholar]