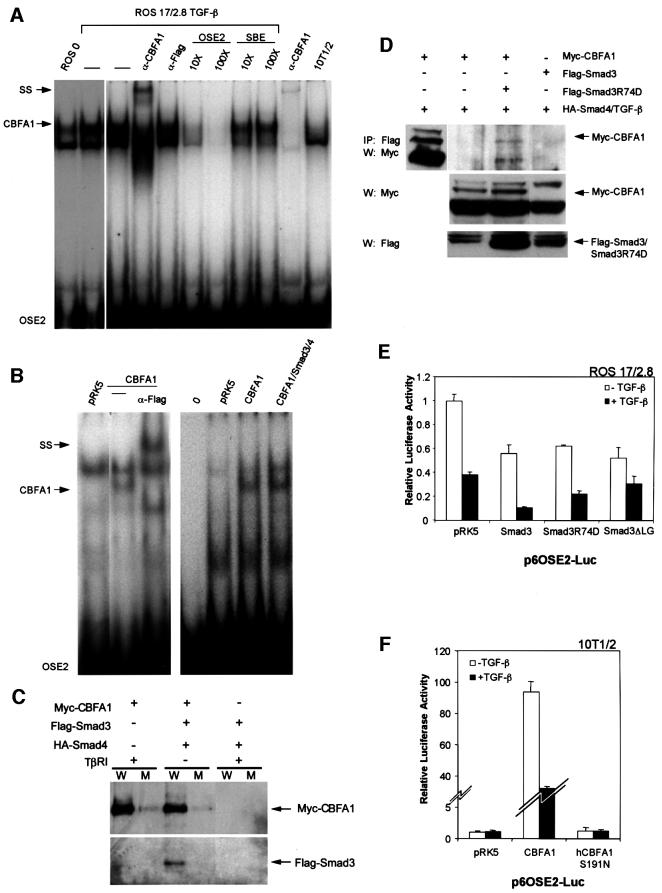
Fig. 8. Binding of CBFA1 and Smad3 to the OSE2 DNA sequence. (A) Electrophoretic mobility shift assays using untreated (lane 1) or TGF-β-treated (5 ng/ml; lane 2) ROS 17/2.8 cell extracts allowed visualization of endogenous CBFA1 binding (arrow) to a 32P-labeled OSE2 oligonucleotide. The identity of the CBFA1-containing complex was confirmed by its absence in 10T1/2 cells, which lack CBFA1 expression (lane 10), and its partial disappearance and supershift (SS) following incubation with an anti-CBFA1 antibody (α-CBFA1, lane 4), but not with an unrelated antibody (α-Flag, lane 5). Unlabeled OSE2 competitor oligonucleotide, at 10- to 100-fold molar excess, competed (lanes 6 and 7), whereas an unrelated competitor oligonucleotide did not compete (lanes 8 and 9) for the radiolabeled OSE2–CBFA1 complex. (B) Electrophoretic mobility shift assays show CBFA1 binding to an OSE2 oligonucleotide, both in the absence or presence of Smad3 and Smad4. COS-1 cells were transfected with expression plasmids for Flag-CBFA1, with or without Smad3/4 (as shown) or the pRK5 control plasmid. The radiolabeled OSE2 oligonucleotide was incubated alone (0) or in the presence of cell lysates. Expression of Flag-CBFA1 resulted in the formation of a distinct band (lanes 2 and 6), which could be supershifted (SS) using an anti-Flag antibody (lane 3). This OSE2–CBFA1 complex was also apparent in the presence of overexpressed Smad3/4 (lane 7). (C) Binding of CBFA1 to an OSE2 oligonucleotide, in the absence or presence of equimolar levels of Smad3 and Smad4. Biotinylated wild-type (W) or mutant (M) OSE2 oligonucleotides were incubated with the same lysates as in Figure 7C, and the interactions of Myc-CBFA1 and Flag-Smad3 with the oligonucleotides were assessed by western blotting. CBFA1 binds to the wild-type oligonucleotide, both in the presence or absence of Smad3/4, but not to the mutant oligonucleotide. Furthermore, Smad3 interacts with the OSE2 nucleotide only in the presence of CBFA1. (D) The DNA binding-defective Smad3R74D interacts with CBFA1 in transfected 10T1/2 cells. Immunoprecipitation assays, performed as in Figure 7C and D, revealed coprecipitation of Myc-CBFA1 (arrow) with Flag-Smad3R74D (top panel). Western analysis without prior immunoprecipitation visualized the migration of Myc-CBFA1 (top panel, left lane), and the expression levels of CBFA1 and Smad3 or Smad3 R74D (lower panels). (E) DNA binding of Smad3 is not essential for TGF-β/Smad3-mediated repression of transcription from the OSE2 sequence by CBFA1. ROS 17/2.8 cells were transfected with the p6OSE2-Luc reporter and wild-type or mutant Smad3 expression plasmids. Luciferase expression in the absence or presence of TGF-β(1 ng/ml) was scored as in Figure 2. (F) DNA binding of CBFA1 is essential for transcriptional activity and Smad3-mediated repression. 10T1/2 cells were transfected with the p6OSE2-Luc reporter and an expression plasmid for wild-type CBFA1 or the DNA binding-defective mutant hCBFA1 S191N. Luciferase expression in the absence or presence of TGF-β (1 ng/ml) was scored as in Figure 2.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.