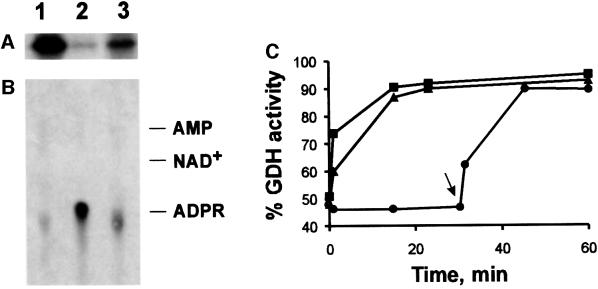
Fig. 8. Reversal of ADP-ribosylation and concomitant reactivation of GDH catalyzed by a mitochondrial hydrolase activity. (A and B) Purified GDH was [32P]ADP-ribosylated and then freed of non-covalently bound nucleotide by HPLC as described in the legend to Figure 5. The modified enzyme (10 µg) was then incubated in the presence of 5 mM MgCl2 (lanes 1), 5 mM MgCl2 and 10 µg of the mitochondrial preparation (lanes 2), or 10 µg of the mitochondrial preparation alone (lanes 3). After 30 min at 30°C, the protein was subjected to SDS–PAGE and the remaining modification visualized by autoradiography (A). The liberated nucleotides were analyzed by thin-layer chromatography (B). The autoradiogram is shown. On the right, the migration of standard compounds is indicated (ADPR, ADP-ribose). A control sample (no addition during incubation) was indistinguishable from the sample represented by lanes 1. (C) Purified GDH or endogenous GDH of the mitochondrial preparation was ADP-ribosylated to achieve ∼50% inhibition by incubating mitochondria and purified GDH (squares and circles) or mitochondria alone (triangles) in the presence of 100 µM NAD+ for ∼45 min (cf. Figure 4). The sample represented by the circles was then centrifuged to remove the added mitochondria. Thereafter (time zero in this figure), 5 mM MgCl2 was added to all samples and incubation continued. At the times indicated, aliquots received 10 mM EDTA and their GDH activity was determined. The arrow indicates re-addition of mitochondria (3 µg per µg of GDH, after 30 min of incubation) to the sample previously freed of mitochondria. The values given are related to the activities of purified or endogenous GDH, respectively, prior to the incubation with NAD+. The data represent 2–4 independent experiments using two different mitochondrial preparations.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.