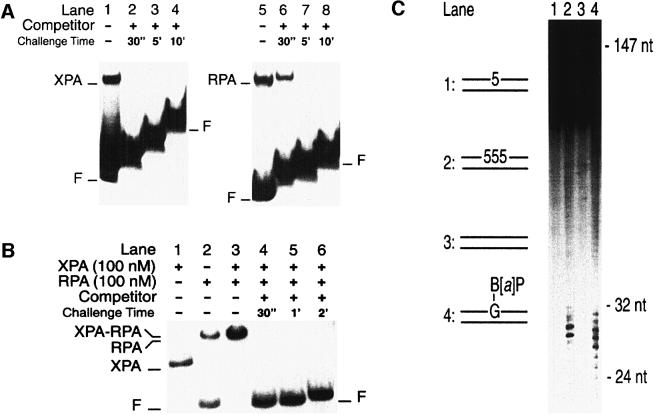
Fig. 7. XPA–RPA generates dynamic complexes with NER substrates. (A) Nucleoprotein complexes were produced by incubating XPA protein (400 nM; lanes 1–4) or RPA (50 nM; lanes 5–8) with 19mer substrates containing a benzo[a]pyrene-dG adduct. Lanes 1 and 5 show the complexes formed during the first 10 min of incubation. After this pre-incubation time, the reactions were supplemented with a 100-fold excess of cold competitor DNA (19mer duplexes with three 5-nitroindole analogues in the centre). The samples were further incubated for the time intervals indicated and loaded on to the gel for electrophoresis. The stepwise appearance of the bands reflects the delay with which the samples were applied on to the running gel. (B) Nucleoprotein complexes produced by incubating XPA and RPA (100 nM each) with 43mer substrates containing three consecutive 5-nitroindoles in the centre. The band shifts formed by either factor alone are shown in lanes 1 (XPA) and 2 (RPA). Lane 3 demonstrates that, together, XPA and RPA bind all the DNA substrate during a pre-incubation of 10 min. The samples of lanes 4–6 were loaded after the time periods indicated, following the addition of a 100-fold excess of competitor DNA. (C) Excision assay in HeLa cell extract. The human NER system excises three consecutive 5-nitroindoles (lane 2). A single 5-nitroindole (lane 1) or undamaged DNA (lane 3) is not repaired. Lane 4 shows a control reaction with substrate containing a benzo[a]pyrene-dG adduct, confirming that the excision products are within the expected size range of 24–32 nucleotides.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.