Abstract
Background
Although the SARS-Cov-2 pandemic has passed, the virus continues to cause infection and death. Immunological response is crucial to overcome the disease. Elucidating the prognostic value of measurable immunological variables at the time of admission and throughout the disease progression could provide stratification tools, improving the management of patients.
Methods
A total of 1,096 samples belonging to 979 hospitalised patients molecularly diagnosed with SARS-CoV-2 infection, together with their medical records were retrospectively analysed. Demographic data, including age, sex, date of symptom onset and admission and T, B and NK cells and serum cytokine levels, were collected as predictor variables. Stratification of patients was performed according to their severity (grade 1: uncomplicated disease; grade 2: mild pneumonia; grade 3: severe pneumonia), their admission to Intensive Care Unit and their final outcome (death vs. alive).
Results
On admission, more severe patients were older, had lower lymphocyte subpopulation counts and higher levels of IL-6, IL-8, IL-10 and IP-10. The trend of lymphocyte counts throughout the disease progression since the symptom onset was to increase both in less and severe patients (even though in these last the levels remained significantly lower), except for NK cells, which decreased. Regarding cytokine levels, IL-6 and IL-8 tended to increase, whereas IL-10 and IP-10 tended to decrease in more severe patients, although particularly IL-10 remained significantly higher in them as compared with less severe patients, and, interestingly, its tendency was to increase in those who died. When considering the clinical evolution, an increase of IL-8 serum levels and a decrease of NK cells were significantly associated with a worsening, while an increase of CD19+ B and CD8+ T cells and a decrease of IL-6, IL-10, MCP-1 and IP-10 cytokine circulating levels were significantly related to an improvement. By integrating all the results, 13 optimal classification trees were constructed using evolutionary algorithms to predict COVID-19 outcome.
Conclusions
The age and the study of lymphocyte populations in a county hospital, together with cytokine serum level quantitation in a third-level hospital, are enough variables to predict the outcome of patients hospitalised due to SARS-CoV-2 infection.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12979-025-00532-w.
Keywords: Severe acute respiratory syndrome coronavirus 2, COVID-19, Immunosenescence, Immunity, Cytokines, Lymphocyte populations
Background
More than five years after the start of the SARS-CoV-2 pandemic, normalcy has apparently prevailed worldwide. However, despite the lower virulence of the majority strains at present, the virus continues to cause infection and death, mainly in at-risk populations. A simple search on the PubMed portal of the National Library of Medicine provides an overwhelming number of publications; more than 481,640 since the beginning of the pandemic and more than 23,271 in the first six months of 2025 [1]. Interest in the infection and related pathology has not waned, and the generation of knowledge for clinical decision-making and resource allocation in the event of a new upsurge of the disease is particularly useful.
For the resolution of the infection, the immune response is essential. This antiviral response begins with the recognition of molecular structures by a whole set of receptors expressed on cells of innate immunity called pattern recognition receptors. Viral pathogen-associated molecular patterns, such as the single-stranded RNA characteristic of coronaviruses, can be detected by PRRs such as RIG-I, MDA5 or TLR-7 and TLR-8. Thus, SARS-CoV-2 infection, activating the TLR pathway, would cause a release of soluble mediators such as proinflammatory Interleukin (IL)−1β and IL-6. Cytokine production acts as a link between the innate and adaptive responses. Among the adaptive responses triggered, the differentiation to Th1 lymphocyte subpopulations with the production of IFN-γ and IL-2, the activation of cytotoxic T-lymphocytes or the production of specific antibodies by B-lymphocytes could be highlighted [2].
However, the immune response itself could exacerbate the course of the disease. Since the first months of the pandemic, the role of both innate and adaptive immunity in the pathogenesis of the infection and its severity became evident on multiple levels: the generation of a hyper-inflammatory state, defects in the production of interferons I and III, lymphopenia or defects of the neutrophil's activation [3]. Studies on lymphocyte populations and cytokine quantification in patients with COVID-19 are abundant because of the critical role that these elements play in the immune response to the virus [4–6]. Severe cases of COVID-19 are often associated with lymphopenia [7], a disbalance in effector T-cells and an increase in activated or exhausted T-cells and a decrease in regulatory T-cells [5]. Other works have explored the prognostic value of lymphocyte subpopulations in predicting disease severity and prognosis in patients with COVID-19 [7, 8]. Moreover, cytokine storm, characterised by excessive and deregulated cytokine production, is a hallmark of severe cases of COVID-19 [9]. Most of the studies have been performed in specialised hospitals where a wide range of diagnostic techniques are available. However, the entry point to the health system is often through regional hospitals. The health systems of our neighbouring countries ensure equity in access to health care. The SARS-CoV-2 pandemic called into question not only the response capacity of these health systems but also the equity of access to them. The availability of reliable evidence in the early stages of the disease is useful to allocate resources correctly and efficiently so that their availability is guaranteed; this premise is applicable in any situation, but even more in those scenarios in which the avalanche of possible needs exceeds the available resources.
Elucidating the prognostic value of immunological parameters at the time of admission may help in decision-making in the management of infection, opening the possibility of identifying biomarkers that allow a more precise stratification of risk groups. In this context, this study aimed to analyse the immune responses in patients hospitalised by COVID-19. Considering that the availability of resources is not homogeneous, the purpose was to provide tools in the form of algorithms, applicable to the hospital management of patients and to early therapeutic decision-making in different healthcare access contexts.
Methods
Aim, design and setting of the study
This study aimed to analyse the immune responses in patients hospitalised in a third level centre by COVID-19 in order to provide tools in the form of algorithms, applicable to the hospital management of patients and to early therapeutic decision-making in different health care access contexts. For this purpose, a retrospective study based on the medical records of patients with confirmed SARS-CoV-2 infection who required hospital admission was conducted. All consecutive patients admitted for this cause at the Reina Sofía University Hospital, Córdoba (third level hospital), from the first week of March 2020 to the first week of June 2021, and who were requested for lymphocyte population studies or quantification of inflammatory cytokines, were included.
Data collection
All data were extracted from electronic medical records. Demographic data (age, sex, date of symptom onset, date of admission, time elapsed from symptom onset to admission) and immunological parameters: counts of lymphocyte populations (T, B and NK cells; cells*10^3/µL), and cytokines, IL-1β, IL-6, IL-8, IL-10, IL-12p70, TNF-ɑ, IFN-γ, MCP-1 and IP-10 (pg/mL), were collected as predictor variables. From 979 patients, a total of 1,096 samples were obtained: 888 at admission (single measurements), and additional samples from repeated measurements in 91 patients during follow-up.
Patients were classified into four groups according to the time elapsed from symptom onset to admission. Thus, those patients whose symptoms started 1 week before their hospitalisation, were grouped as week 1, while those whose symptoms started 4 before their hospitalisation, were grouped as week 4. As outcome variables, severity was stratified according to the criteria of the technical document for the clinical management of COVID-19 in hospital care of the Ministry of Health [10]: 1) Uncomplicated disease; 2) Mild pneumonia, radiographically confirmed. Without signs of severity and with room air SaO2 > 90%; 3) Severe pneumonia, respiratory distress, sepsis, septic shock, or death. The analysis was also performed considering the Intensive Care Unit (ICU) admission and death as outcome variables.
Laboratory procedure
Lymphocyte population counts (cells*10^3/µL), were analysed by flow cytometry (BD Multitest™ 6-color TBNK with BD Trucount™ tubes; ref. 337,166, Becton Dickinson and Company, San Jose, CA, USA). Serum cytokines were quantified by Cytometry Bead Array (CBA, BD Cytometric Bead Array Human Soluble Protein Master, ref. 558,264; Becton Dickinson and Company, San Jose, CA, USA). The following cytokines were analysed according to the manufacturer's instructions: IL-1β (ref. 558,279), IL-6 (ref. 558,276), IL-8 (CXCL8, ref. 558,277), IL-10 (ref. 558,274), IL-12p70 (ref. 558,283), MCP-1 (CCL2, ref. 558,287) TNF-ɑ (ref. 558,273), IFN-γ (ref. 558,269) and IP-10 (CXCL10, ref. 558,280). Samples were acquired in a BD FacsCanto II cytometer, using BD FacsCanto™ as acquisition and analysis software of lymphocyte populations and BD FacsDiva™ software for acquisition and FCAP Array™ software for analysis of cytokine concentrations. A minimum of 500 events were recorded per each cytokine. Median Fluorescence Intensity (MFI) data were transformed into concentration (pg/mL) using a calibration curve as a reference.
Statistical analysis
Demographic and clinical characteristics of patients were described as frequencies or means with standard deviations (SD), according to degree of severity, exitus state or the need of being hospitalised in ICU. Across these outcomes, qualitative variables were compared using the Chi-square test, and quantitative variables were compared using ANOVA (equal or unequal variances) when normality was met (assessed with Shapiro-Wilks test if sample size < 50 and Kolmogorov–Smirnov test for ≥ 50), or Kruskal–Wallis test if not.
The same approach was used to assess whether quantitative variables differed globally over time, as well as at weeks 2, 3, and 4 in comparison to the samples of patients admitted on week 1 after symptom onset. To evaluate trends over time, a p-value for trend was provided. Results from the longitudinal analyses were presented using mean and standard error plots, as well as heatmaps. In the heatmaps, variables significant at 95% or 90% confidence level were shown. A red/orange colour scale was associated with a worse healthy state, whereas green colour scale indicated an improvement on health. The direction of arrows revealed if the value grew (↑) or decreased (↓) compared to week 1. Since only 91 patients (9,3%) were followed for more than one specific week, the statistical analyses were conducted from an independent-groups perspective.
To discover which variables were associated with the outcomes and their interactions, several globally optimal classification trees were learned by using evolutionary algorithms. They worked as follows: a set of trees was initialised with random split rules in root nodes; their structure was modified with mutation and crossover operators applying tests in internal nodes; after each modification, the best candidate models were selected for the next iteration and process ended when the quality of the best tree did not improve further, or the maximum number of iterations was reached. Parameters were fixed on: maximum depth of 7, 100 iterations, 500 trees in the population, a value of alpha equal to 0.05 to regulate the complexity part of the cost function and a numeric seed of 1,000 to initialise the random number generator for reproducibility. These trees were obtained separately in each week for the binary outcomes: degree of severity, exitus state and ICU admission. Due to the presence of missing values, different sets of tentative predictors were trained to ensure an acceptable number of sample size. The criteria followed in selecting the trees for each of the scenarios were the total ‘n’ (representing the number of patients used to construct the model after eliminating patients with missing values for any of the variables loaded), the area under the curve (AUC), the accuracy, and the positive and negative predictive values. Furthermore, in addition to the goodness-of-fit measures, the branches of each tree were also considered, so that the choice would make clinical sense. The decision trees provided probabilities on the final nodes and the cut-off probability to classify a patient with absence or presence of the outcome was defined using the Youden index, which maximised the mean of sensitivity and specificity. Therefore, when the predicted probability exceeded the Youden index cut-off, the patient was classified as having the outcome and was represented by a red circle at the final node. Conversely, if the probability was equal to or below the cut-off, the patient was classified as not having the outcome and was represented by a green circle. According to this classification, true positives and negatives referred to the number of patients correctly classified as having or not having the outcome, respectively, while false positives/negatives represented incorrect classifications. Based on these frequencies, metrics derived from the classification matrix (accuracy, sensitivity, specificity, and predictive values) and AUC were supplied.
The database was structured in SPSS [11] and analysed in R environment [12]. Data were imported from SPSS to R using the “haven” package [13]. Classification trees were obtained with “evtree” [14]. The AUC was computed using the “pROC” package [15].
Results
Demographical and immunological characteristics on admission
A total of 1,096 samples belonging to 979 inpatients at Reina Sofía University Hospital (Córdoba, Spain) with molecularly confirmed SARS-CoV-2 infection were included. According to disease severity, mild cases accounted for 3.8%, moderate cases for 46.5% and severe cases for 49.7%. Of the total, 239 patients (24.4%) were admitted to ICU and 81 patients (8.3%) died. Descriptive baseline characteristics of the population (valid sample size, frequencies, percentages, mean, and SD) as well as categorical variables stratified by severity are shown in Table 1. Sex distribution showed males accounting for 61.2% of cases. Age in our cohort ranged from 18 to 95 years, with a mean of 56.0 years (SD 12.9). The age (p < 0.001) and male sex (p = 0.026) arose as severity determinants in our series.
Table 1.
Demographic characteristics of the study group
| Severity |
Total (n = 979) |
p-value | |||
|---|---|---|---|---|---|
|
Grade 1 (n = 37) |
Grade 2 (n = 455) |
Grade 3 (n = 487) |
|||
| Age, mean (SD) | 51.2 (11.9) | 52.9 (12.3) | 59.2 (12.8) | 56.0 (12.9) | < 0.001 |
| Female genre, n (%) | 22 (59.5) | 187 (41.1) | 171 (35.1) | 380 (38.8) | 0.026 |
| Days since the symptom onset, mean (SD) | 10.5 (5.3) | 9.9 (4.1) | 10.9 (5.8) | 10.4 (5.1) | 0.395 |
| Intensive Care Unit admission, n (%) | 0 | 0 | 239 (49.1) | 239 (24.4) | < 0.001 |
| Death patients, n (%) | 0 | 0 | 81 (16.6) | 81 (8.3) | < 0.001 |
To compare immunological variables with disease severity, the latter was stratified into three levels: 1-for mild cases, 2-for moderates and 3-for severe. Table 2 shows the cross-sectional comparison of each of the predictive variables analysed between patients with severity 3 with respect to those with severity 1 and 2. Likewise, Table 3 presents a cross-sectional comparison between patients who were admitted to the ICU and those who were not, while Table 4 compares patients who died with those who survived.
Table 2.
Predictive variables of the study group according to their severity grade
| Variables on admission | No. grade 1 and 2 | No. grade 3 | Mean (SD) Grade 1 and 2 |
Mean (SD) Grade 3 |
Grade 1 and 2 vs. 3 p-value |
|---|---|---|---|---|---|
| Age (years) | 513 | 583 | 52.7 (12.4) | 59.0 (12.6) | < 0.001 |
| Lymphocyte populations | |||||
| Lymphocytes CD3+/µL | 495 | 537 | 653.1 (421.1) | 546.0 (357.3) | < 0.001 |
| Lymphocytes CD3+CD4+/µL | 495 | 537 | 414.7 (288.3) | 355.0 (251.4) | < 0.001 |
| Lymphocytes CD3+CD8+/µL | 495 | 537 | 225.0 (179.0) | 180.2 (139.8) | < 0.001 |
| CD4/CD8 ratio | 495 | 537 | 2.2 (1.3) | 2.5 (1.6) | 0.006 |
| Lymphocytes CD19+/µL | 464 | 510 | 131.3 (89.2) | 155.6 (372.3) | 0.646 |
| Lymphocytes CD3−CD16+CD56+/µL | 464 | 510 | 173.0 (107.3) | 139.4 (103.3) | < 0.001 |
| Cytokine releasing | |||||
| IL-1β (pg/mL) | 367 | 405 | 0.3 (1.1) | 0.7 (3.3) | 0.812 |
| IL-6 (pg/mL) | 417 | 447 | 20.8 (71.5) | 141.9 (595.9) | < 0.001 |
| IL-8 (pg/mL) | 417 | 447 | 36.5 (47.6) | 100.7 (310.4) | < 0.001 |
| IL-10 (pg/mL) | 417 | 446 | 1.8 (6.8) | 5.9 (14.6) | < 0.001 |
| IL-12p70 (pg/mL) | 348 | 393 | 0 (0.1) | 0.3 (2.8) | 0.135 |
| IFN-γ (pg/mL) | 351 | 390 | 0.2 (2.3) | 0.2 (1.2) | 0.781 |
| TNF-α (pg/mL) | 368 | 403 | 0.1 (0.8) | 0.8 (6.5) | 0.076 |
| MCP-1 (pg/mL) | 121 | 122 | 167.5 (242.2) | 277.6 (383.4) | < 0.001 |
| IP-10 (pg/mL) | 121 | 122 | 1348.2 (1134.6) | 2986.7 (3625.3) | < 0.001 |
Table 3.
Predictive variables of the study group according to their ICU admission
| Variables on admission | No. no ICU admission | No. ICU admission | Mean (SD) No ICU admission |
Mean (SD) ICU admission |
No vs. ICU admission p-value |
|---|---|---|---|---|---|
| Age (years) | 779 | 317 | 54.7 (13.1) | 59.3 (11.8) | < 0.001 |
| Lymphocyte populations | |||||
| Lymphocytes CD3+/µL | 747 | 285 | 615.0 (403.4) | 551.2 (359.5) | 0.014 |
| Lymphocytes CD3+CD4+/µL | 747 | 285 | 392.1 (278.7) | 361.3 (249.6) | 0.127 |
| Lymphocytes CD3+CD8+/µL | 747 | 285 | 210.4 (165.9) | 178.9 (146.4) | < 0.001 |
| CD4/CD8 ratio | 747 | 285 | 2.21 (1.4) | 2.6 (1.7) | < 0.001 |
| Lymphocytes CD19+/µL | 704 | 270 | 128.6 (89.2) | 184.3 (503.4) | 0.094 |
| Lymphocytes CD3−CD16+CD56+/µL | 704 | 270 | 172.3 (108.1) | 111.2 (88.4) | < 0.001 |
| Cytokine releasing | |||||
| IL-1β (pg/mL) | 549 | 223 | 0.4 (1.1) | 0.8 (4.3) | 0.437 |
| IL-6 (pg/mL) | 618 | 246 | 27.9 (101.5) | 222.9 (784.1) | < 0.001 |
| IL-8 (pg/mL) | 618 | 246 | 41.5 (61.0) | 140.8 (408.0) | < 0.001 |
| IL-10 (pg/mL) | 618 | 245 | 2.8 (9.5) | 6.8 (15.5) | < 0.001 |
| IL-12p70 (pg/mL) | 525 | 216 | 0.1 (1.2) | 0.3 (3.4) | 0.814 |
| IFN-γ (pg/mL) | 525 | 216 | 0.2 (2.1) | 0.1 (0.8) | 0.541 |
| TNF-α (pg/mL) | 546 | 225 | 0.2 (1.2) | 1.3 (8.5) | 0.045 |
| MCP-1 (pg/mL) | 177 | 66 | 175.6 (238.8) | 349.2 (465.3) | < 0.001 |
| IP-10 (pg/mL) | 177 | 66 | 1589.0 (1831.9) | 3731.3 (4107.4) | < 0.001 |
Table 4.
Predictive variables of the study group according to their outcome
| Variables on admission | No. alive | No. deceased | Mean (SD) alive |
Mean (SD) deceased |
Alive vs. deceased p-value |
|---|---|---|---|---|---|
| Age (years) | 989 | 107 | 54.9 (12.4) | 66.8 (12.8) | < 0.001 |
| Lymphocyte populations | |||||
| Lymphocytes CD3+/µL | 936 | 96 | 608.7 (396.1) | 486.8 (339.8) | < 0.001 |
| Lymphocytes CD3+CD4+/µL | 936 | 96 | 389.9 (272.8) | 322.6 (249.1) | 0.004 |
| Lymphocytes CD3+CD8+/µL | 936 | 96 | 206.2 (164.1) | 157.4 (123.0) | < 0.001 |
| CD4/CD8 ratio | 936 | 96 | 2.3 (1.4) | 2.8 (2.0) | 0.023 |
| Lymphocytes CD19+/µL | 881 | 93 | 136.5 (105.5) | 215.9 (834.3) | 0.020 |
| Lymphocytes CD3−CD16+CD56+/µL | 881 | 93 | 159.6 (104.7) | 115.2 (115.1) | < 0.001 |
| Cytokine releasing | |||||
| IL-1β (pg/mL) | 705 | 67 | 0.5 (2.5) | 0.4 (2.4) | 0.060 |
| IL-6 (pg/mL) | 792 | 72 | 65.5 (348.7) | 280.4 (952.6) | < 0.001 |
| IL-8 (pg/mL) | 792 | 72 | 58.4 (169.5) | 194.2 (542.2) | < 0.001 |
| IL-10 (pg/mL) | 792 | 71 | 3.7 (11.7) | 6.6 (11.2) | 0.005 |
| IL-12p70 (pg/mL) | 677 | 64 | 0.2 (2.2) | 0 (0) | 0.451 |
| IFN-γ (pg/mL) | 676 | 65 | 0.2 (1.8) | 0.2 (1.2) | 0.719 |
| TNF-α (pg/mL) | 705 | 66 | 0.5 (4.8) | 0.5 (3.9) | 0.667 |
| MCP-1 (pg/mL) | 231 | 12 | 202.4 (299.4) | 615.1 (522.6) | < 0.001 |
| IP-10 (pg/mL) | 231 | 12 | 2032.6 (2703.8) | 4831.6 (3530.6) | < 0.001 |
According to severity, the age was significantly higher in patients with severity 3 regarding those with severity 1–2 (59.0 SD 12.6 vs. 52.7 SD 12.4; p < 0.001). In addition, more severe patients exhibited significantly lower counts of total T CD3+ lymphocytes (546 SD 357.3 vs. 653.1 SD 421.1; p < 0.001), T CD4+ (355.0 SD 251.4 vs. 414.7 SD 288.3; p < 0.001), T CD8+ (180.2 SD 139.8 vs. 225.0 SD 179.0; p < 0.001) and NK cells (139.4 SD 103.3 vs. 173.0 SD 107.3; p < 0.001) as compared with those with severity 1–2, but significantly increased serum levels of IL-6 (141.9 SD 595.9 vs. 20.8 SD 71.5; p < 0.001), IL-8 (100.7 SD 310.4 vs. 36.5 SD 47.6; p < 0.001), IL-10 (5.9 SD 14.6 vs. 1.8 SD 6.8; p < 0.001), MCP-1 (277.6 SD 383.4 vs. 167.5 SD 242.2; p < 0.001) and IP-10 (2986.7 SD 3625.3 vs. 1348.2 SD 1134.6; p < 0.001) cytokines. The CD4/CD8 ratio was significantly higher in more severe patients (2.5 SD 1.6 vs. 2.2 SD 1.3; p = 0.006), probably due to a more pronounce decrease in T CD8+ cells regarding the decrease of T CD4+ cells in them (Table 2).
When considering ICU admission (Table 3), those patients who were admitted to ICU during their hospitalisation stay had lower counts of total T CD3+ lymphocytes (551.2 SD 359.5 vs. 615.0 SD 403.4; p = 0.014), T CD8+ (178.9 SD 146.4 vs. 210.4 SD 165.9; p < 0.001) and NK cells (111.2 SD 88.4 vs. 172.3 SD 108.1; p < 0.001) and significantly increased serum levels of IL-6 (222.9 SD 784.1 vs. 27.9 SD 101.5; p < 0.001), IL-8 (140.8 SD 408.0 vs. 41.5 SD 61.0; p < 0.001), IL-10 (6.8 SD 15.5 vs. 2.8 SD 9.5; p < 0.001), MCP-1 (349.2 SD 465.3 vs. 175.6 SD 238.8 p < 0.001) and IP-10 (3731.3 SD 4107.4 vs. 1589.0 SD 1831.9; p < 0.001) cytokines. The CD4/CD8 ratio was also significantly increased in those patients who were admitted to ICU (2.6 SD 1.7 vs. 2.2 SD 1.4; p < 0.001), again probably due to a more pronounce decrease in T CD8+ cells regarding the decrease of T CD4+ cells in these patients.
Finally, taking into account the final outcome (Table 4), the age (66.8 SD 12.8 vs. 54.9 SD 12.4; p < 0.001), total T CD3+ lymphocytes (486.8 SD 339.8 vs. 608.7 SD 396.1; p < 0.001), T CD4+ (322.6 SD 249.1 vs. 389.9 SD 272.8; p = 0.004), T CD8+ (157.4 SD 123.0 vs. 206.2 SD 164.1; p < 0.001), B CD19+ (315.9 SD 834.3 vs. 136.5 SD 105.5; p = 0.020) and NK cells (115.2 SD 115.1 vs. 159.6 SD 104.7; p < 0.001), as well as serum levels of IL-6 (280.4 SD 952.6 vs. 65.5 SD 348.7; p < 0.001), IL-8 (194.2 SD 542.2 vs. 58.4 SD 169.5; p < 0.001), IL-10 (6.6 SD 11.2 vs. 3.7 SD 11.7; p = 0.005), MCP-1 (615.1 SD 522.6 vs. 202.4 SD 299.4; p < 0.001) and IP-10 (4831.6 SD 3530.6 vs. 2032.6 SD 2702.8; p < 0.001) cytokines showed significant differences between deceased and alive patients. Once more, the CD4/CD8 ratio was also significantly higher in those patients who finally died (2.8 SD 2.0 vs. 2.3 SD 1.4; p = 0.023).
The age
Analysis of the behaviour of the age variable showed that all patients who were hospitalized later during the curse of the illness were older, with significant differences in the most severe group as well as in those with mild and moderate severity. Therefore, the age again behaves as a key factor in COVID-19 disease progression. Thus, it was significantly higher in patients with severity 3 than those with severity 1 and 2 at week 3 (62.7 SD 10.4 vs. 53.0 SD 13.0) and, indeed, patients having a delayed hospital admission were older, both for severity 1–2 (p-trend = 0.026) and severity 3 (p-trend = 0.011; Supplementary Table 1). The mean age of patients admitted to the ICU was also significantly higher as their hospitalisation delayed, particularly in weeks 2 (59.1 SD 11.5 vs. 55.5 SD 13.1) and 3 (62.6 SD 10.1 vs. 55.5 SD 13.1; p-trend = 0.004; Supplementary Table 2). Concerning the group of patients who died (Supplementary Table 3), the mean age was higher overall in each of the weeks of admission, with p-trend < 0.001 in favour of those who survived.
Analysis of lymphocyte populations
Variations in circulating lymphocyte populations considering the moment of hospital admission since the disease symptom onset with respect to week 1 were presented in Fig. 1 and Supplementary Tables 1–3, together with the trend over time (p-trend). In all cases, the analysis showed the overall results and the data for each week of hospital admission.
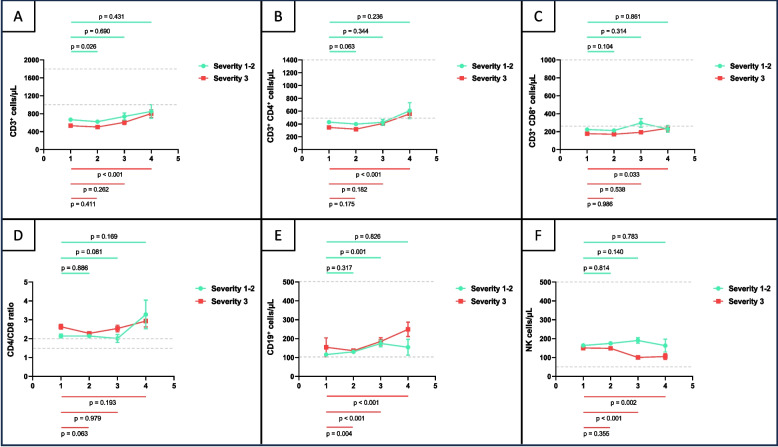
Fig. 1.
Lymphocyte subpopulation counts (T and B lymphocytes and NK cells) by Flow cytometry (No./µL) and CD4+/CD8+ ratio (A-F) in the study group in each week post symptom onset (week-1 to week-4), according to the disease severity (grade 1–2 vs. 3). Mean and standard error of the mean, together with comparisons regarding baseline (week 1) are displayed. Grey bars indicate normal ranges
Total T CD3+ cells were significantly lower in week 2 as compared with week 1 in patients with severity 1–2 (p = 0.026), but significantly increased in week 4 as compared with week 1 in patients with severity 3 (p < 0.001; Fig. 1.A). T CD4+ (p < 0.001) and T CD8+ (p = 0.033) cells also appeared significantly increased in week 4 regarding week 1 in more severe patients (Fig. 1.B and C). Conversely, NK cells significantly decreased in those patients in weeks 3 (p < 0.001) and 4 (p = 0.002) regarding week 1 (Fig. 1.F). Concerning CD19+ B cells, they appeared significantly increased in week 3 regarding week 1 (p = 0.001) in patients with severity 1–2 and in week 2 (p = 0.004), week 3 (p < 0.001) and week 4 (p < 0.001) as compared with week 1 in patients with severity 3 (Fig. 1.E). No statistically significant variations were found regarding the remaining periods of time in lymphocyte subpopulation circulating levels in each group of severity.
The trend of total lymphocytes and CD4+ T cells seemed to be similar, improving over time, with significant recovery (p-trend < 0.001 in both cases) observed in patients with severity 3 and those admitted to ICU (p < 0.001 in both cases; Supplementary Table 2). Surviving patients (Supplementary Table 3) also showed a positive trend in these parameters (CD3+ p-trend = 0.007; CD4+ p-trend = 0.007). CD8+ lymphocytes showed a significant change over time in patients who were admitted to ICU (p-trend = 0.019; Supplementary Table 2) and those who survived (p-trend = 0.026; Supplementary Table 3). NK cells remained stable in patients with severity 1–2. However, in patients with severity 3 and those who died, they were significantly lower over time (p-trend < 0.001; p-trend = 0.033; Supplementary Table 3). The trend of CD19+ lymphocytes was to increase throughout time in all severity groups (Supplementary Table 1), but with a significant p-trend in less severe patients (p-trend < 0.001), those who were not admitted to ICU (p-trend < 0.001) and who did not die (p-trend < 0.001; Supplementary Tables 1–3).
Analysis of cytokine releasing
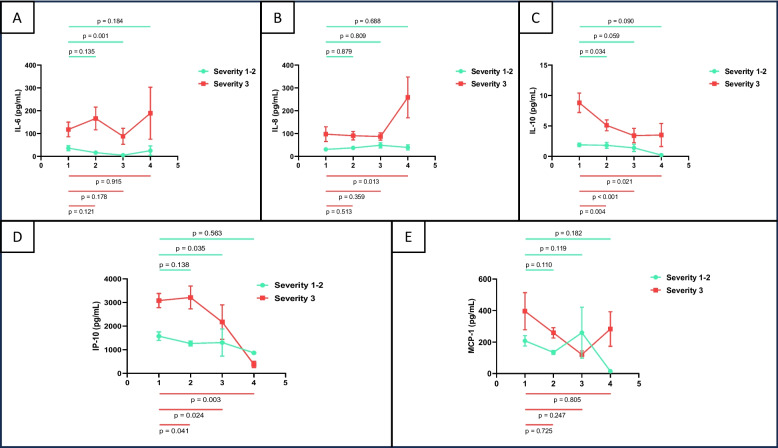
Variations of the different cytokines are shown in Fig. 2, as well as in Supplementary Tables 1–3. IL-6 (p = 0.001) and IP-10 (p = 0.035) serum levels from patients with severity 1–2 significantly decreased in week 3 regarding week 1 (Fig. 2.A and D). IL-10 also significantly decreased in this group of patients, but in week 2 regarding week 1 (p = 0.034; Fig. 2.C). When analysing cytokine serum levels of more severe patients, we found that IL-8 significantly increased in week 4 regarding week 1 (p = 0.013; Fig. 2.B) and that IL-10 and IP-10 significantly decreased as the time elapsed from the symptom onset to hospitalisation (Fig. 2.C and D). No statistically significant variations were found regarding the remaining periods of time in cytokine serum levels in each group of severity. As a general rule, the technique used has not been proven sensitive enough for detecting IL-1β, IL-12p70, IFN-γ and TNF-⍺ (Tables 2–4).
Fig. 2.
Serum cytokine (IL-6, IL-8, IL-10, MCP-1 and IP-10) levels (pg/mL) by flow cytometry (Cytometric Bead Array) in the study group (A-E) in each week post symptom onset (week-1 to week-4), according to the disease severity (grade 1–2 vs. 3). Mean and standard error of the mean, together with comparisons regarding baseline (week 1) are displayed
When considering the whole evolution, the trend of IL-6 was to significantly decrease in less severe patients (p-trend = 0.016; Supplementary Table 1). The trend of IL-10 was also to significantly decrease but in more severe ones (p-trend = 0.007; Supplementary Table 1). However, both cytokine levels remained higher in more severe patients regarding less ones. This trend was also observed when comparing those patients who were admitted to ICU with those who were not (IL-6 p-trend = 0.049 no-ICU; IL-10 p-trend = 0.013 ICU; Supplementary Table 2). Interestingly, serum IL-10 levels tended to significantly decrease in patients who remained alive (p-trend = 0.001; Supplementary Table 3). In contrast, even though no significantly, they tended to increase in those who died (p-trend = 0.527; Supplementary Table 3), being higher at all time-points than that from those from alive patients.
Significant variables related to outcome
The Fig. 3 displays how immunological variables changed throughout disease progression and whether these variations were associated with a significant improvement or worsening. Considering all studied variables, the decrease in IL-10, MCP-1, IP-10 and IL-6 serum levels generally was associated with a significant improvement for both severity, ICU admission and final outcome. An improvement of disease progression was also associated with an increase in T CD8+ and B CD19+ cells with hospital admission time. In contrast, the decrease in NK cell peripheral counts and the increase in IL-8 serum levels were significantly associated with a worsening of the disease. The changes in the remaining variables could be associated with either an improvement or a worsening according to the considered outcome (severity, ICU admission or death) at each time (weeks 1–4). Even though an apparent increase in IL-1ꞵ and a decrease in IFN-γ levels seemed to be related to a worsening and an improvement of disease progression, respectively, the absence of sensitivity in the detection test used, exclude them as immunological determinant biomarkers, suitable for the development of the decision algorithms.
Fig. 3.
Heatmap longitudinal-analysis plot including the immunological variables as biomarkers of disease progression. The red/orange colour scale was associated with worsening, whereas the green colour scale, with improving. Darkness of the colour is proportional to statistical significance. Direction of arrows indicates whether the variable increases (↑) or decreases (↓) regarding week 1 considering the outcome in each case (severity of the disease, Intensive Care Unit -ICU- admission or final outcome)
Dynamic decision trees
By integrating all the results, optimal classification trees were constructed using evolutionary algorithms. These trees were obtained separately for each week for the binary outcomes: severity, ICU admission and death. For each case, those best suited to the laboratory resources usually available at different levels of care, either in a county hospital or in a third level hospital, were chosen. Each tree was named using the letter S for severity, I for ICU admission and D for death; this was followed by the week number 1 to 3 from symptom onset to hospital admission; finally, the letter C for county hospitals or T for third level hospitals was added. In several cases where the same tree was appropriate for both types of hospitals, the letters C_T were entered. For simplicity, the diagnostic performance statistics for each tree is shown in Table 5. The figures for each of the 13 selected trees are shown as Fig. 4. The estimated probability of the outcome is displayed at the final node. A red circle indicates that the patient was classified as having the outcome, and a green circle indicates classification as not having the outcome. For example, in the first week, the algorithm S1C had the best diagnostic performance to discriminate severity in a county hospital and was based on the values of lymphocyte populations and sex (Fig. 4 and Table 5). In the case of the third-level hospital, the S1T tree uses the values of IL-8, IL-10, IL-6, total T-lymphocytes and CD8+ T-lymphocytes to obtain the best diagnostic performance (Fig. 4 and Table 5).
Table 5.
Statistical parameters obtained in each selected decision tree
| S1C | S1T | S2C | S2T | S3C_T | I1C | I1T | I2C | I2T | I3C_T | D1C_T | D2C_T | D3C_T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of samples included | 288 | 199 | 518 | 139 | 127 | 288 | 188 | 518 | 139 | 127 | 288 | 351 | 127 |
| True positives | 122 | 83 | 132 | 58 | 74 | 36 | 30 | 48 | 22 | 42 | 15 | 8 | 9 |
| False negatives | 38 | 27 | 111 | 13 | 4 | 38 | 18 | 71 | 8 | 11 | 19 | 14 | 8 |
| False positives | 37 | 16 | 44 | 13 | 12 | 12 | 8 | 22 | 4 | 5 | 7 | 4 | 2 |
| True negatives | 91 | 73 | 231 | 55 | 37 | 202 | 132 | 377 | 105 | 69 | 247 | 325 | 108 |
| Prevalence | 0.556 | 0.553 | 0.469 | 0.511 | 0.614 | 0.257 | 0.255 | 0.230 | 0.216 | 0.417 | 0.118 | 0.063 | 0.134 |
| Accuracy | 0.74 | 0.784 | 0.701 | 0.813 | 0.874 | 0.826 | 0.862 | 0.820 | 0.914 | 0.874 | 0.910 | 0.949 | 0.921 |
| Sensitivity | 0.763 | 0.755 | 0.543 | 0.817 | 0.949 | 0.486 | 0.625 | 0.403 | 0.733 | 0.792 | 0.441 | 0.364 | 0.529 |
| Specificity | 0.711 | 0.820 | 0.840 | 0.809 | 0.755 | 0.944 | 0.943 | 0.945 | 0.963 | 0.932 | 0.972 | 0.988 | 0.982 |
| Positive predictive value | 0.767 | 0.838 | 0.750 | 0.817 | 0.860 | 0.750 | 0.789 | 0.686 | 0.846 | 0.894 | 0.682 | 0.667 | 0.818 |
| Negative predictive value | 0.705 | 0.73 | 0.675 | 0.809 | 0.902 | 0.842 | 0.88 | 0.842 | 0.929 | 0.863 | 0.929 | 0.959 | 0.931 |
| Area under the curve | 0.783 | 0.833 | 0.725 | 0.854 | 0.881 | 0.772 | 0.803 | 0.68 | 0.862 | 0.895 | 0.725 | 0.683 | 0.757 |
| Cut-off | 0.389 | 0.320 | 0.440 | 0.313 | 0.286 | 0.250 | 0.150 | 0.164 | 0.075 | 0.250 | 0.077 | 0.042 | 0.069 |
Fig. 4.
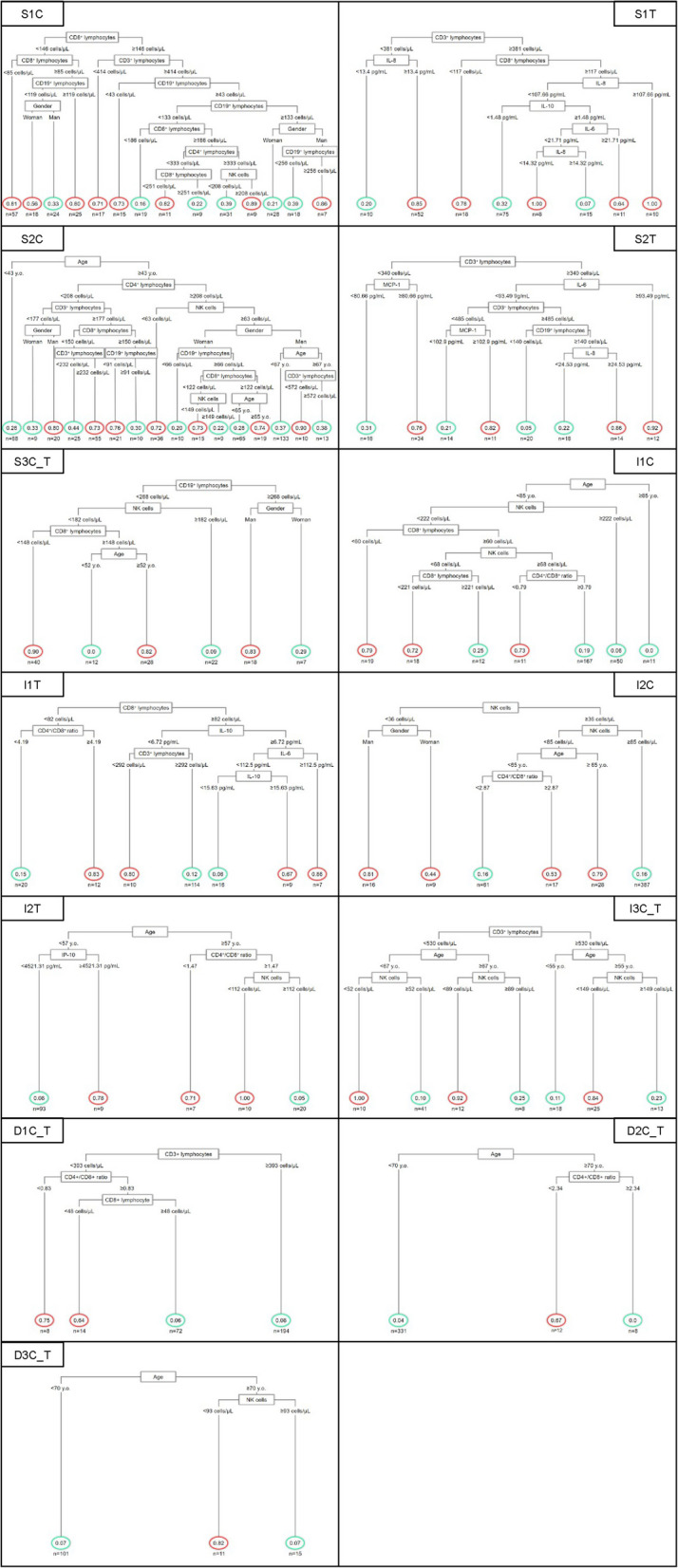
Algorithm classification trees for disease severity integrating the age, sex, lymphocyte subpopulation counts and serum cytokine levels throughout hospitalisation time for the binary outcomes: severity, Intensive Care Unit (ICU) admission and death, at different levels of care, either in a county or a third-level hospital. Each tree is named using the letter S for severity, I for ICU admission and D for death, followed by the week number 1–3 and the letter C for county and T for third-level hospitals. At the final node, patients classified as having the outcome are indicated with a red circle, while those without the outcome are indicated with a green circle
Discussion
Over the more than five years since the pandemic began, numerous studies have investigated the role of the immune system in disease pathogenesis, as well as the utility of specific immunological biomarkers for predicting patient outcomes and monitoring treatment responses. Having reliable indicators available in the early stages of the disease could support the appropriate and efficient allocation of resources, ensuring their availability. This principle holds true in any scenario but becomes even more critical when the surge in potential needs outpaces available resources. Most studies examining the role of immunological biomarkers in predicting the course of COVID-19 have been performed in specialised hospitals, where a broad range of diagnostic techniques is available. However, initial access to healthcare typically occurs through regional hospitals. The healthcare systems of our neighbouring countries aim to ensure equitable access to care, but the SARS-CoV-2 pandemic challenged not only their response capacity but also the equity of access. This research shows that the determination of basic immunological parameters, such as the quantification of lymphocyte populations, allows applying decision trees to characterise the risk of severity, hospitalisation in ICU or death at different times of evolution in patients with COVID-19. Where cytokine quantification is available, as would be the case in tertiary hospitals, the predictive capacity of decision trees can be improved, although in the third week of admission, lymphocyte counts are sufficient in all cases.
In the early months of the pandemic, patient classification systems emerged that attempted to predict risk for severity based on clinical and analytical parameters. Since the seminal works, older age, male sex, the existence of certain comorbidities and elevated acute phase reactants have been associated with a worse prognosis [16–18]. Among the laboratory parameters, lymphocyte counts, and the quantification of pro-inflammatory cytokines have figured prominently. In the current study, these immune parameters have been analysed cross-sectionally in a cohort of hospitalised patients, taking into account the time elapsed from the symptom onset.
In the cross-sectional design phase of the present study, the value of age, elevated pro-inflammatory cytokines and decreased lymphocyte counts as factors linked to a worse disease outcome was reconfirmed. Years have passed since the first wave of the pandemic, but throughout 2023 and 2024 work has continued to emerge that attempts to predict disease progression based on demographics, clinical symptoms, analytical parameters or imaging tests. Most of these studies had a cross-sectional design based on data obtained at the earliest time of hospitalisation. These include the multicentre research by Guo W et al. [19], using a multivariate logistic regression analysis resulting in a scoring model, in which age and IL-6 were independent predictors of mortality. Similar was the regression model of the study by Zhao et al. [20], which highlighted the value of age as a predictor of fatality. In the studies published in 2024 by Asteris PG et al. and Quian FH et al. [21, 22], cross-sectional predictive models of severity were developed, both of which emphasised the value of the neutrophil/lymphocyte ratio. Also in 2024, Baek S et al. [23], studying data from 5945 patients, developed a deep neural network model for the initial triage with 11 routine laboratory parameters obtained in the first days of hospitalisation. The model showed an AUC with excellent discriminative ability (AUROC = 0.937). In the 2024 article by Engoren et al. [24], data from 6741 patients were analysed. Initially, a first logistic regression model was developed to assess the risk of disease severity with clinical and laboratory parameters which was later modified with the inclusion of new patients. Although a learning-by-doing approach, it was still a cross-sectional one. Other recent research aimed at predicting disease progression had focused on immunological parameters. Herrera-Van Oostdam AS et al. [25], conducted a cross-sectional study with a panel of immunological parameters, including kynurenine/tryptophan ratio and IL-6, which supplied a high area under the curve (AUC (95%CI) of 0.991 (0.986–0.995) to predict the risk of sepsis and sepsis mortality in patients with COVID-19. Similarly, He X et al. [26], conducted a cross-sectional study in which immunological parameters obtained from mass cytometry by time of flight were used to develop a predictive criticality model based on machine learning.
Notwithstanding the predictive power of each of these models individually, their partial design has been questioned by a Cochrane meta-analysis in 2024, which included the results of 64 studies involving 71,170 patients, evaluating the power of laboratory parameters to predict severe COVID-19 and mortality [7]. Even though the neutrophil-to-lymphocyte ratio increase, and the lymphocyte count decrease were among the five-laboratory data with the highest predictive value, none of the parameters analysed had enough discriminative power to discard a severe disease. Therefore, the authors concluded that an enrichment with other variables such as clinical data or imaging studies should be needed to improve the severity predictive ability of the model.
Based on a longitudinal design, there were far fewer studies that propose algorithms to help predict the evolution of patients hospitalised for COVID-19. In the study by Lourenço AA et al. [27]. an algorithm was developed using laboratory data obtained at three points during hospitalisation. The authors point out that the importance of each of these parameters varies throughout the hospitalisation, emphasising the importance of a dynamic approach. Chen X et al. [28], conducted a longitudinal study in which 14 laboratory parameters were assessed at three time points during hospitalisation. The overall model obtained showed good discriminative performance between patients who survived and those who died (mean AUCs of 88.81). Likewise, Diray-Arce J et al. [29], performed an interesting work with multi-omics data obtained from blood, serum, plasma, and nasal exudate samples and using techniques such as CyTOF, GWAS and Olink. Based on deep immunophenotyping data from more than 15,000 samples obtained from 540 patients, this study defined five cellular and molecular patterns at hospitalisation and longitudinally, capable of distinguishing mild to fatal COVID-19. Unfortunately, this interesting work is hardly replicable in a regional hospital. But even in a tertiary level hospital, it would be difficult to obtain these data in an appropriate time frame to be able to apply clinical decisions and allocate resources.
In contrast to the referenced studies, the main novelty of the present work lies in the analysis of the temporal evolution of these variables and the proposal of decision-making trees adapted to the resources available in a county hospital and in a third-level hospital. These trees are tailored to the week of symptom onset, cover each of the subsequent four weeks, and account for each possible clinical scenarios.
Therefore, the patients who were hospitalised later, whatever the severity of their condition, had a higher mean age, confirming the influence of the age as a key factor in the outcome of the patients. The rising T-lymphocytes, especially the CD4+ count, was a marker of favourable progression over time. A reduction of B-lymphocytes was a sign of a worse outcome. Similarly, NK cells declined progressively in the most severe patients or those who passed away. With respect to the assessment of the cytokines, the most revealing patterns associated with a favourable outcome were the decrease of IL-6, IL-10, MCP-1 and IP-10.
The main limitation of the study was that since it was retrospective, it was not possible to obtain all the data required at each point in the evolution of the disease. Likewise, asymptomatic patients or those whose severity did not require hospitalisation were not included in the study population.
Conclusions
In conclusion, among the immunological study variables, an increase of IL-8 serum levels and a decrease of NK cells were significantly associated with a worsening of the COVID-19 disease progression. In contrast, an increase of CD19+ B and CD8+ T cells and a decrease of IL-6, IL-10, MCP-1 and IP-10 cytokine circulating levels were significantly related to an improvement. Therefore, the age and the study of lymphocyte populations in a county hospital, together with cytokine serum level quantitation in a third-level hospital are shown as enough variables to predict the outcome of patients hospitalised due to SARS-CoV-2 infection, regarding severity, ICU admission and death. Furthermore, the approach proposed by us could be quickly extrapolated to other infective contexts or, more interestingly, to future possible pandemic situations.
Supplementary Information
Acknowledgements
The authors thank all caregivers who indirectly contributed to this multicentre study. Particularly, the authors thank the technical support of Francisco García Hens and María José Morales Serrano.
Abbreviations
- ICU
Intensive Care Unit
- SD
Standard Deviation
Authors’ contributions
AJ and JM conceived the idea for this study, designed the protocol. AN, AT-A, PA, AC, RQ, LC-C, JMV, CF, JM y AJ, collected the data and assessed for data quality. MM-A analysed the data. PA, AJ, JM, and AN drafted the manuscript, but all the authors provided critical revisions and approved the final version of the manuscript.
Funding
This work has been carried out without funding.
Data availability
All collected data, including fully anonymised participant data, are available to others. Available information includes fully anonymised participant data and data dictionary. Related documents are available from the date of publications henceforth: study protocol, statistical analysis, and approval of the ethical board. These documents are available from the date of publications henceforth at email address jeduar.molina.sspa@juntadeandalucia.es Data will be shared after approval of proposals by the Reina Sofía Hospital Ethical Committee.
Declarations
Ethics approval and consent to participate
The study was subject to the standards of good clinical practice and always compiled with the ethical principles contained in the Declaration of Helsinki and subsequent amendments and the basic Law 41/2002 regulating patient autonomy and the rights and obligations regarding clinical information and documentation. The study was approved by the Research Ethics Committee of the Hospital Reina Sofía de Córdoba (ref. 4888). The nature of the study did not imply the performance of any intervention on the patients in addition to the usual care practice, but only reviewed the clinical data collected in their medical records, which were treated anonymously. For this purpose, patients were assigned an alphanumeric code and the identification was kept in the hospital to ensure data confidentiality. All data were processed in accordance with the European Parliament Regulation 2016/679 of April 27, 2016, in force in May 2018 for the Protection of Personal Data.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Paula Álvarez and Ana Navas have equally contributed and share first authorship position.
References
- 1.National Library of Medicine. PubMed search: (COVID-19) OR (SARS-CoV-2). Bethesda (MD): National Center for Biotechnology Information; ]. Available from: https://pubmed.ncbi.nlm.nih.gov/?term=%28covid-19%29+OR+%28SARS-CoV-2%29&sort=date&size=200. Cited 2025 Jun 30.
- 2.Liu WJ, Zhao M, Liu K, Xu K, Wong G, Tan W, et al. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naiditch H, Betts MR, Larman HB, Levi M, Rosenberg AZ. Immunologic and inflammatory consequences of SARS-CoV-2 infection and its implications in renal disease. Front Immunol. 2025;15:1376654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snopkowska Lesniak SW, Maschio D, Henriquez-Camacho C, Moreno Cuerda V. Biomarkers for SARS-CoV-2 infection. A narrative review. Front Med (Lausanne). 2025;12: 1563998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiti L, Markovič T, Lainscak M, Farkaš Lainščak J, Pal E, Mlinarič-Raščan I. The immunopathogenesis of a cytokine storm: the key mechanisms underlying severe COVID-19. Cytokine Growth Factor Rev. 2025;82:1–17. [DOI] [PubMed] [Google Scholar]
- 6.Andrejkovits ÁV, Huțanu A, Manu DR, Dobreanu M, Văsieșiu AM. Dynamic changes in lymphocyte populations and their relationship with disease severity and outcome in COVID-19. Int J Mol Sci. 2024;25(22): 11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rop L, Bos DA, Stegeman I, Holtman G, Ochodo EA, Spijker R, et al. Cochrane COVID-19 Diagnostic Test Accuracy Group; Verbakel JY. Accuracy of routine laboratory tests to predict mortality and deterioration to severe or critical COVID-19 in people with SARS-CoV-2. Cochrane Database Syst Rev. 2024;8(8): CD015050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedayati-Ch M, Ebrahim-Saraie HS, Bakhshi A. Clinical and immunological comparison of COVID-19 disease between critical and non-critical courses: a systematic review and meta-analysis. Front Immunol. 2024;15:1341168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Astroth C, Shah KS, Agrawal S, Agrawal A. Weathering the storm: how age and biologics influence the COVID-19 cytokine surge. Pathogens. 2025;14(4):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spanish Ministry of Health. Clinical management protocol for patients with COVID-19 disease (version of 28 March 2022). Madrid: Government of Spain; 2022. Available from: https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Protocolo_manejo_clinico_ah_COVID-19.pdf. Cited 2025 Jun 30.
- 11.SPSS, “Data Analysis Software System, Version 30,” 2006. Available from: https://www.spss.com.
- 12.R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: https://www.R-project.org/.
- 13.Wickham H, Miller E, Smith D (2023). _haven: Import and Export 'SPSS', 'Stata' and 'SAS' Files. R package version 2.5.4, Available from: https://CRAN.R-project.org/package=haven.
- 14.Thomas Grubinger, Achim Zeileis, Karl-Peter Pfeiffer. evtree: Evolutionary Learning of Globally Optimal Classification and Regression Trees in R. J Stat Softw. 2014;61(1), 1–29. Available from: http://www.jstatsoft.org/v61/i01/.
- 15.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. Available from: http://www.biomedcentral.com/1471-2105/12/77/ [DOI] [PMC free article] [PubMed]
- 16.Bentivegna M, Hulme C, Ebell MH. Primary care relevant risk factors for adverse outcomes in patients with COVID-19 infection: a systematic review. J Am Board Fam Med. 2021;34(Suppl):S113–26. [DOI] [PubMed] [Google Scholar]
- 17.Jurado A, Martín MC, Abad-Molina C, Orduña A, Martínez A, Ocaña E, et al. COVID-19: age, interleukin-6, C-reactive protein, and lymphocytes as key clues from a multicentre retrospective study. Immun Ageing. 2020;17:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín MC, Jurado A, Abad-Molina C, Orduña A, Yarce O, Navas AM, et al. The age again in the eye of the COVID-19 storm: evidence-based decision making. Immun Ageing. 2021;18(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W, Li X, Ding C, Dai X, Wu S, Shi Y, et al. Development and validation of a scoring system to predict the mortality of hospitalized patients with SARS-CoV-2 Omicron: a nationwide, multicentre study. BMC Pulm Med. 2024;24(1):312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao A, Liu Y, Xia J, Huang L, Lu Q, Tang Q, et al. Establishment and validation of a prognostic model based on common laboratory indicators for SARS-CoV-2 infection in Chinese population. Ann Med. 2024;56(1): 2400312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asteris PG, Gandomi AH, Armaghani DJ, Kokoris S, Papandreadi AT, Roumelioti A, et al. Prognosis of COVID-19 severity using DERGA, a novel machine learning algorithm. Eur J Intern Med. 2024;125:67–73. [DOI] [PubMed] [Google Scholar]
- 22.Qian FH, Cao Y, Liu YX, Huang J, Zhu RH. A predictive model to explore risk factors for severe COVID-19. Sci Rep. 2024;14(1):18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baek S, Jeong YJ, Kim YH, Kim JY, Kim JH, Kim EY, et al. Development and validation of a robust and interpretable early triaging support system for patients hospitalized with COVID-19: predictive algorithm modeling and interpretation study. J Med Internet Res. 2024;26: e52134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engoren M, Pancaro C, Yeldo NS, Kerzabi LS, Douville N. Comparison of static and rolling logistic regression models on predicting invasive mechanical ventilation or death from COVID-19-a retrospective, multicentre study. Clin Respir J. 2023;17(1):40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera-Van Oostdam AS, Castañeda-Delgado JE, Oropeza-Valdez JJ, Borrego JC, Monárrez-Espino J, et al. Immunometabolic signatures predict risk of progression to sepsis in COVID-19. PLoS ONE. 2021;16(8): e0256784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Cui X, Zhao Z, Wu R, Zhang Q, Xue L, et al. A generalizable and easy-to-use COVID-19 stratification model for the next pandemic via immune-phenotyping and machine learning. Front Immunol. 2024;15:1372539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lourenço AA, Amaral PHR, Paim AAO, Marques-Ferreira G, Gomes-de-Pontes L, da Mata CPSM, da Fonseca FG, Pérez JCG, Coelho-Dos-Reis JGA. Algorithms for predicting COVID outcome using ready-to-use laboratorial and clinical data. Front Public Health. 2024;12: 1347334. 10.3389/fpubh.2024.1347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Gao W, Li J, You D, Yu Z, Zhang M, Shao F, Wei Y, Zhang R, Lange T, Wang Q, Chen F, Lu X, Zhao Y. A predictive paradigm for COVID-19 prognosis based on the longitudinal measure of biomarkers. Brief Bioinform. 2021;22(6): bbab206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diray-Arce J, Fourati S, Doni Jayavelu N, Patel R, Maguire C, Chang AC, et al. Multi-omic longitudinal study reveals immune correlates of clinical course among hospitalized COVID-19 patients. Cell Rep Med. 2023;4(6): 101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All collected data, including fully anonymised participant data, are available to others. Available information includes fully anonymised participant data and data dictionary. Related documents are available from the date of publications henceforth: study protocol, statistical analysis, and approval of the ethical board. These documents are available from the date of publications henceforth at email address jeduar.molina.sspa@juntadeandalucia.es Data will be shared after approval of proposals by the Reina Sofía Hospital Ethical Committee.