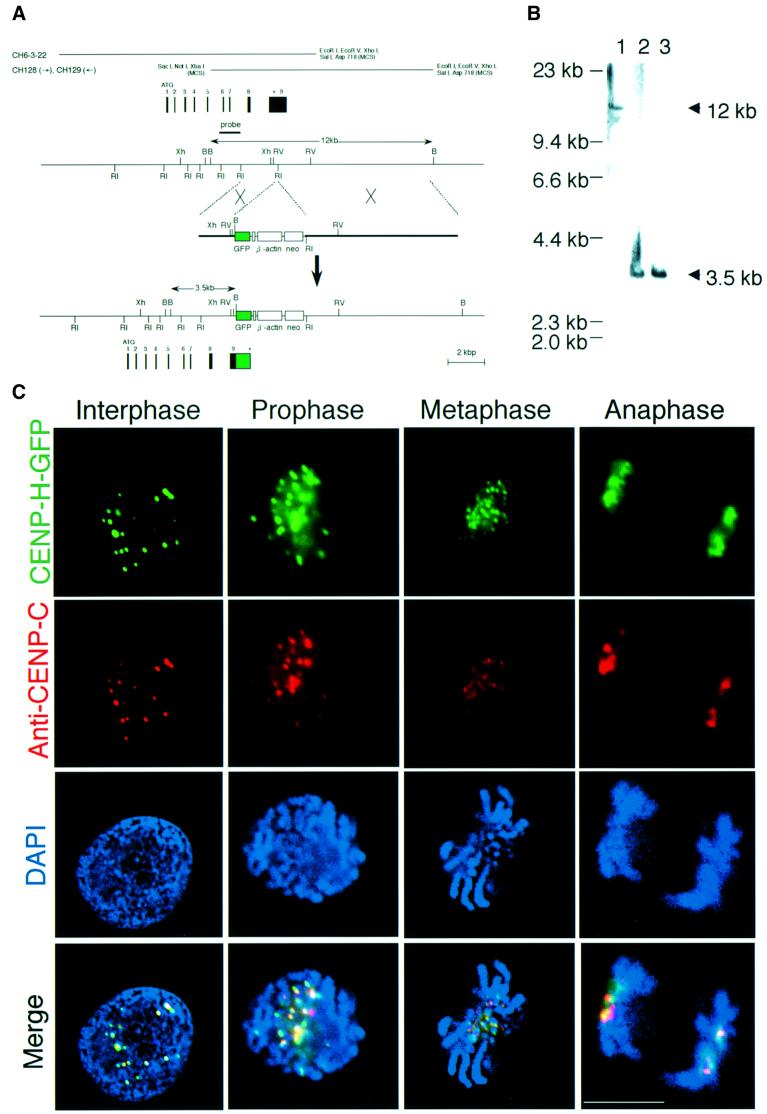
Fig. 2. Chicken CENP-H localizes to centromeres throughout the cell cycle. (A) Restriction maps of the CENP-H gene, the CENP-H–GFP replacement construct and the configuration of the targeted locus. Black boxes indicate the positions of exons. Exon 9, which contains the stop codon, was replaced with a portion of the sequence of exon 9 fused to a GFP tag followed by the neo-resistance gene (neo) and 3′ homology region. Relevant restriction enzyme sites shown are as follows: RI, EcoRI; Xh, XhoI; B, BamHI; RV, EcoRV. The position of the probe used for Southern hybridiza tion is indicated. A new 3.5 kb BamHI fragment hybridizes with the probe if targeted integration of the indicated replacement construct occurs. (B) Restriction analysis of the CENP-H–GFP allele. Genomic DNAs from wild-type DT40 cells (lane 1) and the targeted clones (lanes 2 and 3) were digested with BamHI, size fractionated by 0.7% agarose gel electrophoresis, transferred to a nylon filter and hybridized with the probe indicated in (A). (C) Localization of CENP-H–GFP at progressive stages of the cell cycle in DT40 cells. Cells were fixed and stained with anti-CENP-C antibody (red), and green signals are specific for CENP-H–GFP. Nuclei and chromosomes were visualized by counterstaining with DAPI (blue). The scale bar corresponds to 10 µm. As shown in the merged images, CENP-H–GFP signals are co-localized with CENP-C signals throughout the cell cycle.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.