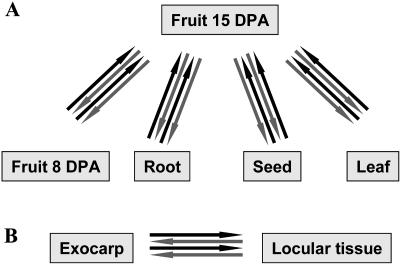
Figure 1.
Design of microarray experiment. A, Comparison of transcript abundance in developing tomato fruit, root, leaf, and seed from cherry tomato (cv WVa106). RNA was extracted from each organ sample (two RNA samples per organ), reverse transcribed, Cy labeled (Cy3 and Cy5 dye-swap for each RNA sample), and hybridized to tomato cDNA arrays for comparison with 15-DPA fruit sample, used as common reference (for each comparison, four slides, i.e. eight subarrays analyzed). B, Comparison of transcript abundance in exocarp and locular tissue from expanding tomato fruit (cv Ferum). Cytological observations and comparison of growth curves indicate that the developmental stage of 22-DPA Ferum fruit is equivalent to that of 15-DPA WVa106 fruit (data not shown). Exocarp and locular tissue samples were obtained by dissecting and pooling fruit tissues from 150 Ferum fruit harvested at 22 DPA. RNA was extracted from pooled tissue samples (two RNA samples per tissue), reverse transcribed, Cy labeled (Cy3 and Cy5 dye-swap for each RNA sample), and hybridized to tomato cDNA arrays (four slides, i.e. eight subarrays analyzed).