Abstract
The proinflammatory cytokine interleukin 17 (IL-17) is the founding member of a family of secreted proteins that elicit potent cellular responses. We report a novel human IL-17 homolog, IL-17F, and show that it is expressed by activated T cells, can stimulate production of other cytokines such as IL-6, IL-8 and granulocyte colony-stimulating factor, and can regulate cartilage matrix turnover. Unexpectedly, the crystal structure of IL-17F reveals that IL-17 family members adopt a monomer fold typical of cystine knot growth factors, despite lacking the disulfide responsible for defining the canonical ‘knot’ structure. IL-17F dimerizes in a parallel manner like neurotrophins, and features an unusually large cavity on its surface. Remarkably, this cavity is located in precisely the same position where nerve growth factor binds its high affinity receptor, TrkA, suggesting further parallels between IL-17s and neurotrophins with respect to receptor recognition.
Keywords: activated T cell/cystine knot/cytokines/IL-17/X-ray crystallography
Introduction
Interleukin 17 (IL-17; hereafter referred to as IL-17A) is a proinflammatory cytokine that is produced by activated T cells (Yao et al., 1995a,b; Fossiez et al., 1996; reviewed in Spriggs, 1997; Lebecque et al., 2001a). IL-17A is secreted as a disulfide-linked homodimeric glycoprotein with a dimer molecular weight of 30–35 kDa (Fossiez et al., 1996). Recently, a number of homologous proteins have been identified; IL-17B, C and E (Li et al., 2000; Shi et al., 2000; Lee et al., 2001) share a conserved C-terminal region, but have N-terminal segments that differ. IL-17B, C and E have significantly different biological profiles from IL-17A. For instance, IL-17B is expressed in pancreas, small intestine and stomach, while IL-17C is very rarely expressed, and none of the three homologs is expressed by activated T cells (Li et al., 2000; Lee et al., 2001). Like IL-17A, the sequences of these proteins are not homologous to other known proteins and thus their mode of receptor interaction can not be predicted without obtaining direct structural information on a family member.
Two homologous receptors for IL-17 family members have been identified, IL-17 receptor (IL-17R) (Yao et al., 1995a, 1997; reviewed in Lebecque et al., 2001b) and IL-17R homolog 1 (IL-17Rh1, also known as IL-17BR) (Shi et al., 2000; Lee et al., 2001). IL-17R is widely expressed and is reported to bind IL-17A with an affinity at least 10-fold weaker (10–50 nM) than the potency of IL-17A on responsive cells, suggesting the possibility that there may be additional components involved in IL-17-induced signaling (Yao et al., 1997). IL-17Rh1 is expressed mostly in liver and kidney tissue and does not bind IL-17A, but instead is reported to bind IL-17B and E (Shi et al., 2000; Lee et al., 2001).
Signaling by IL-17A and IL-17E results in the activation of NF-κB (Yao et al., 1995a; Awane et al., 1999; Lee et al., 2001). Furthermore, IL-17A has been shown to regulate the activities of extracellular regulated kinase (ERK)1, ERK2, c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinases (Shalom-Barak et al., 1998; Awane et al., 1999), with JNK activation depending on the presence of tumor necrosis factor (TNF) receptor associated factor-6 (Schwandner et al., 2000). The proinflammatory function and intracellular signaling pathway of IL-17A are strikingly similar to those of the IL-1 and Toll receptors (reviewed in Bowie and O’Neill, 2000; Daun and Fenton, 2000). Despite this similarity in biological function to other receptor families, the protein sequences of IL-17R and IL-17Rh1 do not resemble those of any known proteins and therefore yield no clues as to their structure and signaling mechanisms.
IL-17A is overexpressed in a number of pathological conditions including inflammatory airway disease (Lindén et al., 2000), transplant rejection (Van Kooten et al., 1998), psoriasis (Teunissen et al., 1998), multiple sclerosis (Matusevicius et al., 1999) and rheumatoid arthritis. In particular, IL-17A is found at high levels in the synovial fluid of patients with inflammatory arthritis and is believed to play a role in the inflammation and skeletal destruction characteristic of this disease (Chabaud et al., 1999; Kotake et al., 1999; Ziolkowska et al., 2000). Accordingly, neutralization of IL-17A in synovial cultures from patients with rheumatoid arthritis causes a substantial reduction in the biological activity of the diseased synovium (Chabaud et al., 1998, 1999; Kotake et al., 1999). IL-17A induces production of, and can synergize with, other proinflammatory cytokines, such as IL-1 and TNF-α (Fossiez et al., 1996; Chabaud et al., 1998, 1999; Jovanovic et al., 1998). Thus, IL-17A may be involved in initiation and maintenance of the proinflammatory cytokine cascade, and, as such, may initiate as well as amplify the skeletal destruction that occurs in patients with inflammatory disorders such as rheumatoid arthritis.
Herein, we report a novel IL-17 family member, IL-17F, the sequence of which is 50% identical to IL-17A. Like IL-17A, IL-17F is secreted from activated T cells, induces cytokine production, and may prove to play an important role in skeletal destruction and inflammation in pathological conditions such as rheumatoid arthritis. While the activity of IL-17F appears to be related to that of IL-17A, the potency differs, consistent with differences in receptor binding affinities. Surprisingly, IL-17F adopts a cystine knot fold and dimerization mode reminiscent of the neurotrophin family of cystine knot growth factors (McDonald et al., 1991). This structure provides a platform from which to compare structural features that will likely be shared among the IL-17 family members, and consider those that must clearly differ. The unexpected similarity between IL-17s and neurotrophins suggests potential mechanisms for receptor binding.
Results
Identification of IL-17F
IL-17A is now recognized as the prototype member of an emerging family of cytokines (Yao et al., 1995a,b; Li et al., 2000; Shi et al., 2000; Lee et al., 2001). Four members of this family have been reported previously and analysis of human genomic sequence indicates the existence of at least two additional members, including the molecule described here, termed IL-17F (Figure 1). The gene encoding human IL-17F is located adjacent to IL-17A (human genomic sequence in clone RP11-935B23; DDBJ/EMBL/Genbank accession No. AL355513). A cDNA corresponding to IL-17F was isolated and found to encode a protein of 163 amino acids, including a 30-residue signal sequence, with the mature protein bearing 50% amino acid identity to IL-17A and identity to a cDNA isolated by Jacobs et al. (1997) and DDBJ/EMBL/Genbank accession No. AF384857. IL-17A and F share a more limited 16–30% amino acid identity with IL-17B, C and E, suggesting that these two form a distinct subgroup within the IL-17 family.
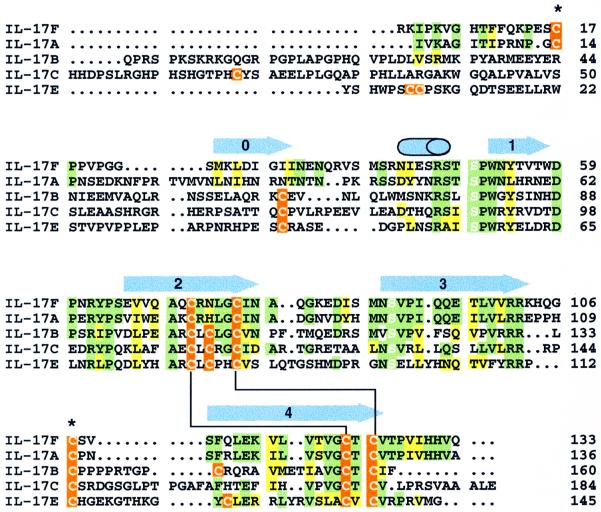
Fig. 1. Sequence alignment of IL-17F with other IL-17 family members. Regions of sequence identity or similarity between IL-17F and IL-17A are highlighted in green and yellow, respectively. When the other family members also have an identical or similar residue at the same position, they are similarly colored. Cysteine residues are indicated in orange. The conserved serines that replace the canonical knot cysteines are highlighted with white letters. Disulfide bonds that are expected to be conserved in all IL-17s are indicated by a black line connecting the bonded cystines. The two cysteines that form the inter-chain disulfide in IL-17F are marked with an asterisk. Secondary structural elements in IL-17F are shown above the sequences as blue arrows (β-strands) or a cylinder (α-helix). Residue numbering is from the start of the mature sequences.
IL-17F mRNA expression
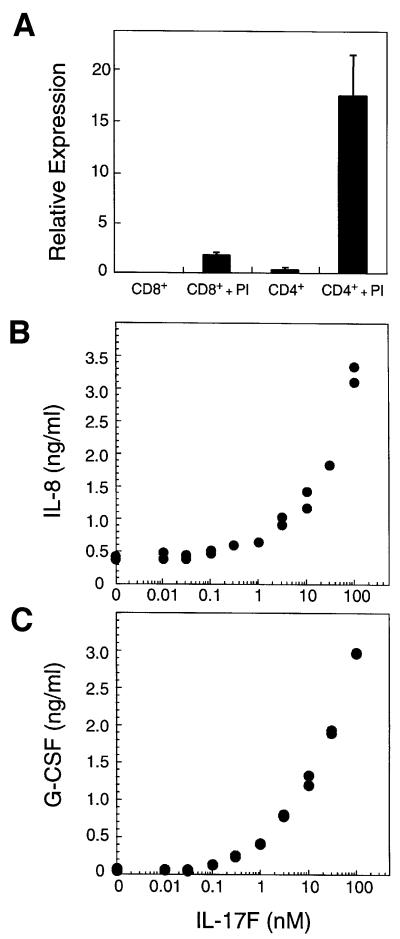
The expression of IL-17F in various human tissues was examined by northern blot analysis. IL-17F transcripts could not be detected in kidney, thymus, testes, trachea, stomach, fetal brain, lung, colon, intestine, prostate, placenta or pancreas. Next, IL-17F mRNA levels were measured in purified T cells. Little message was present in unstimulated CD4+ or CD8+ T cells; however, IL-17F mRNA was dramatically induced in activated CD4+ T cells (Figure 2A). Interestingly, a significant level of expression was also detected in activated CD8+ T cells. Thus, IL-17F expression is rare, but is highly induced in activated CD4+ T cells.
Fig. 2. IL-17F is produced by activated T cells and stimulates cytokine production. (A) IL-17F expression in T cells. Relative mRNA expression is shown. PI, treated with PMA and ionomycin. Induction of IL-8 (B) and G-CSF (C) in fibroblasts by IL-17F. Human primary foreskin fibroblasts were cultured for 24 h in the presence of the indicated concentrations of IL-17F. Conditioned medium was then analyzed by ELISA for the presence of IL-8 and G-CSF.
IL-17F stimulates cytokine production
The sequence similarity between IL-17F and IL-17A as well as their expression within activated T cells raises the possibility that they may possess similar biological activities. We therefore investigated whether IL-17F could induce production of cytokines known to be regulated by IL-17A (Fossiez et al., 1996). Primary human fibroblasts treated with IL-17F exhibited greatly elevated production of IL-8 and granulocyte colony-stimulating factor (G-CSF) in a dose-dependent manner (Figure 2B and C). Similar responses were observed with various cell lines tested (human 293, 3T3 and TK10 cells; not shown), suggesting that IL-17F possesses a broad ability to induce cytokine production.
Effect of IL-17F on cartilage matrix turnover
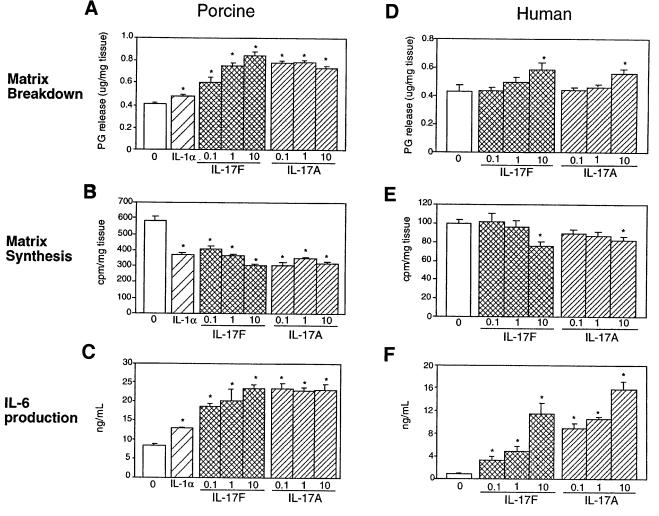
As a potent proinflammatory cytokine produced by activated T cells, IL-17A has been suggested to play a role in inflammatory disorders such as rheumatoid arthritis (reviewed in Lebecque et al., 2001a). To determine whether IL-17F might be capable of mediating similar effects on cartilage matrix metabolism, porcine and human articular cartilage explants were treated with a range of IL-17F concentrations, and proteoglycan release and synthesis were measured. In both systems IL-17F induced significant cartilage matrix release (Figure 3A and D) and inhibited new cartilage matrix synthesis (Figure 3B and E) in a dose-dependent manner. These effects were of the same order of magnitude as that of the known catabolic cytokine, IL-1α. At higher concentrations (10 nM), IL-17F and IL-17A showed equal potency on human articular cartilage matrix turnover (Figure 3D and E). Thus, IL-17F can directly regulate cartilage matrix turnover; however, the potency of IL-17F relative to that of IL-17A depends on the species tested and may relate to receptor affinity.
Fig. 3. Effect of IL-17F on porcine and human cartilage. Porcine articular cartilage (A–C) and human articular cartilage (from 65-year-old caucasian female) (D–F) explants were treated with varying concentrations (0.1, 1 or 10 nM) of IL-17F or IL-17A, or 0.1 nM human IL-1α, and proteoglycan breakdown (A and D), proteoglycan synthesis (B and E) and IL-6 production (C and F) were measured. Data represent the average of five independent samples ± SEM. ‘0’ in each panel represents the untreated control. *denotes statistically significant difference from control (p<0.05).
IL-17A substantially induces expression of IL-6, a potent regulator of bone turnover, and IL-8 in human cartilage, but does not change the levels of IL-2, IL-4, IL-5, interferon-γ or TNF-α (Cai et al., 2001). In both human and porcine articular cartilage, IL-17F could also induce IL-6 production in a dose-dependent manner (Figure 3C and F). In particular, IL-17F, like IL-17A, induced IL-6 at concentrations (0.1 and 1 nM) at which no significant change in matrix turnover or synthesis in human cartilage was observed (Figure 3C and F). In addition, IL-17F was less potent than IL-17A in both porcine and human cartilage in terms of IL-6 production in contrast to the similarity in potency on human cartilage matrix turnover.
Receptor binding
Surface plasmon resonance (SPR) was used to determine whether IL-17F binds the extracellular domains (ECDs) of either of the two receptors reported previously to bind IL-17 proteins. Surprisingly, no binding of either IL-17R or IL-17Rh1 (up to 1 and 0.5 µM, respectively) was observed to immobilized IL-17F. In contrast, IL-17R bound immobilized IL-17A with a modest binding affinity (due to a relatively slow on-rate; Table I), consistent with previous reports on the affinity for this interaction (Yao et al., 1997). Likewise, IL-17Rh1 showed high affinity binding to IL-17E (Table I), consistent with the potency observed for induction of IL-8 release from cells (Lee et al., 2001). As expected (Shi et al., 2000; Lee et al., 2001), no binding was observed between IL-17Rh1 and IL-17A and between IL-17E and IL-17R.
Table I. IL-17 ligand/receptor binding kinetics.
| Immobilized protein | Ligand | Kon × 10–5 (M–1 s–1) | Koff × 104 (s–1) | KD (nM) |
|---|---|---|---|---|
| IL-17A | IL-17R | 0.093 | 6.7 | 72 |
| IL-17E | IL-17Rh1 | 6.7 | 7.0 | 1.1 |
| IL-17Rh1 | IL-17E | 4.3 | 6.2 | 1.4 |
To test whether the lack of IL-17R or IL-17Rh1 binding to IL-17F could be the result of immobilization-linked inactivation, IL-17F/receptor binding was tested in competition experiments. In these experiments a fixed concentration of IL-17R (500 nM) or IL-17Rh1 (31 nM) was incubated with a varied concentration of ligand and then injected over the IL-17A or IL-17E surface. While soluble IL-17A could efficiently block binding of IL-17R to immobilized IL-17A, no competition was observed with 2 µM IL-17F. Furthermore, 1.3 µM IL-17F could not block binding of IL-17Rh1 to immobilized IL-17E, although binding was completely inhibited by soluble IL-17E. These results indicate that IL-17F does not bind with high affinity to the purified, monomeric ECD of either IL-17R or IL-17Rh1.
Structure determination
The structure of human IL-17F was solved by multiwavelength Hg anomalous diffraction methods and was refined to an Rfree and Rcryst of 28.4 and 23.8%, respectively, at 2.85 Å resolution (see Table II and Materials and methods). The core of the IL-17F protomer is composed of two pairs of antiparallel β-strands; one pair includes strands 1 (residues 52–58) and 2 (residues 66–72 and 77–79), while the other includes strands 3 (residues 89–103) and 4 (residues 110–125). Strand 2 is interrupted by a short stretch of irregular β-structure. Two disulfide bridges (Cys72/Cys122 and Cys77/Cys124) connect strands 2 and 4. A third disulfide (Cys17/Cys107) connects the loop between strands 3 and 4 of one protomer to the N-terminal extension of the adjacent monomer forming extensive dimer contacts (see below). This N-terminal extension (residues 1–48) also contains a β-strand (strand 0, residues 25–32), which hydrogen bonds to strand 3′ on the other protomer, and a small α-helix (residues 43–48). Additional electron density was observed at Asn53, consistent with glycosylation of this residue, as was expected from sequence analysis and characterization of the purified protein.
Table II. Crystallographic statistics.
| Data collection and MAD phasing | ||||
|---|---|---|---|---|
| Native | Hg peak | Hg inflection | Hg remote | |
| Space group | P65 | |||
| Unit cell constants (Å) | a = 126.4, c = 89.9 | a = 126.8, c = 90.0 | ||
| Wavelength (Å) | 0.979 | 1.0067 | 1.0087 | 1.127 |
| Max. resolution (Å) | 2.85 | 2.8 | 2.8 | 2.65 |
| Completeness (%)a | 100 (100) | 98.9 (91.3) | 98.8 (90.2) | 99.9 (99.9) |
| Rsymb | 0.088 (0.54) | 0.058 (0.34) | 0.059 (0.36) | 0.064 (0.43) |
| Reflections measuredc | 141 778 | 228 788 | 228 553 | 266 735 |
| Reflections uniquec | 19 294 | 40 450 | 40 459 | 46 851 |
| Phasing power centricd | – | 1.4 | 1.6 | 1.3 |
| Phasing power acentricd | – | 4.3 | 3.2 | 4.3 |
| Cullis R acentricd |
– |
0.76 |
0.7 |
0.8 |
| Refinement | ||||
| Resolution (Å) | 30–2.85 | r.m.s.d. bonds (Å) | 0.012 | |
| No. reflections (F >0.2σ) | 19 246 | r.m.s.d. bonded Bs (Å2) | 0.012 | |
| Re | 0.238 | r.m.s.d. angles (°) | 1.6 | |
| Rfree | 0.284 | No. of protein atoms | 3725 | |
| Residues | No. of carbohydrate atoms | 84 | ||
| chain A | 8–128 | No. of solvent molecules | 28 | |
| chain B | 8–128 | |||
| chain X | 9–18, 22–128 | |||
| chain Y | 6–18, 24–130 | |||
aNumbers in parentheses refer to the highest resolution shell.
bRsym = Σ|I – <I>|/ΣI, where <I> is the average intensity of symmetry-related observations of a unique reflection.
cBijvoet reflections are kept separate in the Hg statistics.
dPhasing statistics are for reflections with F >2σ.
eR = Σ|Fo – Fc|/ΣFo
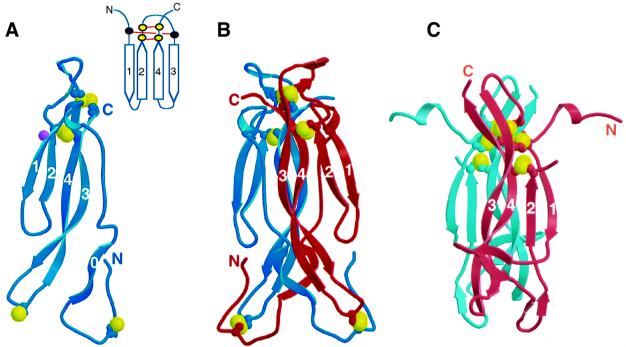
Unexpectedly, this structure reveals that IL-17F is a structural homolog of the cystine knot family of proteins (McDonald and Hendrickson, 1993), named for its unusual cystine connectivity (Figure 4). The cystine knot fold is characterized by two sets of paired β-strands (strands 1 and 2 and strands 3 and 4) that are connected by disulfide linkages between strands 2 and 4 (Figure 4A, inset). A third disulfide bridge passes through this macrocycle to connect strands 1 and 3. In contrast, IL-17F contains only two of the three distinctive cystine linkages that give the family its name. In IL-17F, the Cys72/Cys122 and Cys77/Cys124 disulfides form the macrocycle of the typical cystine knot. The third disulfide that would form the ‘knot’ by passing through this macrocycle is not present; instead, residues 50 and 90, which are located in the same three-dimensional space as the third disulfide in cystine knot proteins, are serines in IL-17F. While Ser50 is in the same χ1 rotamer conformation as the corresponding cysteine in a knot protein, Ser90 is not. Notably, serines are conserved in these positions in all IL-17 family members (Figure 1), despite the fact that the structure suggests the third disulfide could be accommodated.
Fig. 4. The structure of IL-17F. (A) Ribbon trace of the IL-17F monomer. Strands are labeled. Disulfides are shown as ball-and-stick representation with the sulfur atoms colored yellow. Glycosylation of Asn53 is indicated by a purple ball. Inset shows a cartoon representation of the canonical cystine knot fold. Cysteine residues are indicated by filled circles; those present in IL-17 proteins are yellow, whereas the two that are missing are black. (B) Ribbon trace of the IL-17F dimer. Disulfides are shown as in (A). (C) The structure of NGF from the NGF–TrkA complex (Wiesmann et al., 1999; Protein Data Bank code 1WWW); a disordered loop connects strands 2 and 3.
Dimerization
IL-17F dimerizes in a parallel fashion similar to nerve growth factor (NGF) and other neurotrophins (McDonald et al., 1991). However, the dimer interface is unusually large, burying a total of 6800 Å2 (or ∼3400 Å2 per monomer) as compared with 3400 Å2 total (∼1700 Å2 per monomer) for NGF (Wiesmann et al., 1999; Protein Data Bank code 1WWW). Approximately one third of the interface is formed by interactions between strands 3 and 4 of one monomer with the same strands in the other monomer, analogous to the dimer interface seen in neurotrophins. Unique to IL-17F, however, is the vast amount of surface buried by interactions involving the N-terminal extension of each protomer reaching across the canonical dimer interface and packing against various portions of the other protomer.
The overall backbone structure of the IL-17F dimer can be described as a garment where sheets 1/2 and 1′/2′ form the sleeves, the cystine knot disulfides line the collar, and sheets 3/4 and 3′/4′ along with the N-terminal extensions form the body, which is finished off with the two three-stranded sheets (involving strands 4/3/0′ and 0/3′/4′) forming a skirt at the bottom (Figure 4B; dimensions 65 Å × 25 Å × 30 Å). A striking feature on the surface of the molecule is an unusually large cavity (two per dimer; 18 Å × 10 Å × 10 Å) located at the dimer interface essentially positioned as pockets in the garment. The base of the cavity is formed by residues in strands 3 and 3′ (Gln95, Glu96, Thr97 and Leu98 from both chains) and 4 and 4′ (Lys115′, Val118, Val120 and Val120′). Residues in the N-terminus line one side of the cavity (residues Arg37, Val38, Met40), while the other side is lined by residues from strand 1 (Tyr54), strand 2 (Glu66, Val68), and the turn between these strands (Tyr63 and Pro64). The peptide bond between Tyr63 and Pro64 is in the cis conformation. This proline is conserved in all IL-17 sequences and is always preceded by a large hydrophobic residue (Figure 1), suggesting that this peptide bond is likely to be in a cis conformation in all IL-17 family members. The mercury-containing compound, thimerosal, which was used to phase the structure, binds in the lower end of this cavity (as oriented in Figure 5), occupying ∼30% of the space.
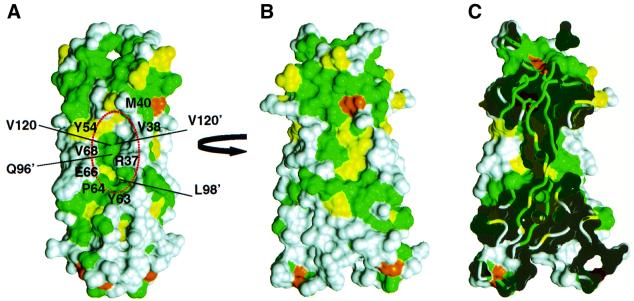
Fig. 5. Comparison of IL-17F and IL-17A. Two orthogonal views, ‘side’ (A) and ‘front’ (B), of the molecular surface of IL-17F colored according to sequence conservation between IL-17A and F as in Figure 1. The surface of residues that are identical between the two proteins are colored green, homologous residues are colored yellow, while residues that differ significantly are colored white. [The view in (B) is oriented ∼15° rotated from the view in Figure 4B.] Residues forming the cavity are labeled in (A). (C) ‘Cut-away’ view of the surface in (B) showing how the large cavities on either side of IL-17F penetrate deeply into the body of the dimer.
Discussion
IL-17F activity
We have identified a novel member of the IL-17 cytokine family, IL-17F. Like IL-17A, IL-17F induces production of other cytokines, chemokines and hematopoietic factors, such as IL-6, IL-8 and G-CSF. IL-17F and IL-17A share similar expression patterns, each apparently restricted to activated T cells (Yao et al., 1995b; Fossiez et al., 1996). However, further investigation may reveal distinct biological roles for IL-17F. The physiological stimuli that lead to the induction of IL-17F expression remain to be determined, and there may be substantial differences in the regulation of IL-17A and F. Interestingly, the gene for IL-17F is located adjacent to IL-17A in the human genome.
The IL-17s constitute an emerging family of cytokines and their associations with human disease have not yet been investigated extensively. However, the potent biological actions that have been observed to date suggest that members of this family likely contribute to immune disorders. Initial reports have pointed to a clear association of IL-17A with rheumatoid arthritis (Chabaud et al., 1999; Kotake et al., 1999; Ziolkowska et al., 2000), a disease characterized by infiltration of leukocytes, synovitis, pannus formation and skeletal destruction (Arend and Dayer, 1990). In humans, activated T cells likely play a key role in the disease process through both direct and indirect mechanisms (Kingsley and Panayi, 1997; Kong et al., 1999; Miossec, 2000). More specifically, activated T cells stimulate other cells such as macrophages and fibroblasts to release cytokines, which can then amplify the local immune response and promote synovitis. Our data showing that IL-17F is also produced by activated T cells and can have direct effects on articular cartilage matrix turnover and IL-6 production in the absence of inflammatory cells suggest that IL-17F may also be able to promote skeletal tissue destruction.
IL-17F appears to have activity related to that of IL-17A, but surprisingly does not bind IL-17R with high affinity in vitro. However, enhanced binding of IL-17F to Cos cells transfected with IL-17R can be detected (data not shown), suggesting that IL-17F may be able to utilize IL-17R, but only in combination with additional, as yet unidentified, components to form a high affinity signaling complex. A similar mechanism has been postulated for IL-17A to explain the >10-fold discrepancy between receptor affinity and the potency of its biological activity (Yao et al., 1997). Our results suggest that regardless of the receptor(s) involved, IL-17F signaling results in similar downstream activities to stimulation of IL-17R by IL-17A.
IL-17s adopt a cystine knot fold
The discovery that an IL-17 homolog is a member of the cystine knot fold superfamily and dimerizes similarly to members of the NGF subfamily was unexpected. IL-17 proteins share negligible sequence similarity with other members of the superfamily. For example, a structure-based sequence alignment of IL-17F with NGF reveals identity for only ten of 133 residues, including the four cysteines conserved in the cystine knot motif (not shown). Limited sequence conservation is typical of the cystine knot fold superfamily (McDonald and Hendrickson, 1993), but the absence of one of the canonical disulfides of the cystine knot is unprecedented. The presence of additional cysteines elsewhere in the structure further confuses arguments about cystine knot family membership based on conserved cysteine spacing (Lebecque et al., 2001a).
The structure of IL-17F allows generalization to the other IL-17 family members. The cystine knot fold including the location of the β-sheets and the macrocycle disulfide linkage should be preserved in all IL-17 homologs (Figure 1). In particular, IL-17A is so similar to IL-17F, with 50% sequence identity, that it is possible to predict where IL-17A and F will share surface features and where they will diverge. Figure 5 shows the molecular surface of IL-17F colored according to sequence identity with IL-17A. The only extensive conserved patches on the surface of IL-17F are on the flat face of each protomer (Figure 5B) and on the area ‘above’ the cavity (Figure 5A). The conserved area on the protomer face may represent either conserved features required for maintaining the structure or for potentially recognizing common binding partners. The large cavity in the surface of IL-17F is also expected to be present in IL-17A, and would be composed of both conserved and variable residues (Figure 5C).
In contrast, the sequences of IL-17B, C and E diverge significantly from IL-17F and A, having longer N-terminal extensions and additional cysteine residues (Figure 1). Furthermore, IL-17B is secreted as a non-covalent dimer (both from CHO or Hi5 cells; Shi et al., 2000; data not shown), indicating that the inter-chain disulfide in IL-17F is not present in IL-17B. In order to accommodate additional or different disulfide linkages in the context of the cystine knot scaffold, IL-17B, C and E are likely to have their N-termini in significantly different conformations than that of IL-17F and A, thus dividing the family into two subclasses. Further structural information about IL-17B, C or E will be necessary to determine precisely how the structure of the N-terminus differs across the family.
Implications for receptor binding
One of the most striking features of the structure of IL-17F is the unusually large cavity formed by residues in the dimer interface (Figure 6A), which is highly suggestive of a region that might bind another molecule. The cavity (two per dimer) is composed of a combination of residues that are either strictly conserved or always possess a similar chemical character (Tyr54, Tyr63, Pro64, Val120), as well as others that are extremely variable among IL-17 family members (Arg37, Val38, Met40, Glu66) providing potential to impart specificity for intermolecular interactions. The cavity does not have a pronounced electrostatic surface feature, but instead is formed by a combination of hydrophobic, polar and charged residues (Figures 5A and 6A). Based on sequence analysis, an analogous cavity would be expected to exist in other IL-17 family members; however, given the likely different conformation of the N-terminal extension, the specific characteristics of the cavity could be quite different in IL-17B, C and E.
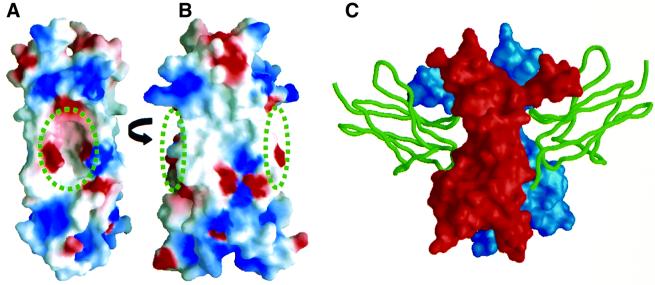
Fig. 6. Comparison of the IL-17F surface and the TrkA-binding site on NGF. (A and B) The molecular surface of IL-17F is oriented as in Figure 5. IL-17F is colored according to the electrostatic surface potential: red, –5 kT; white, 0 kT; and blue, +5 kT. The positions of the cavities are indicated by the circles. (C) The molecular surface of NGF is shown in the same orientation as IL-17F in (B) with the two protomers of the dimer colored red and blue; domain 5 of TrkA is shown as a green ribbon (Wiesmann et al., 1999).
Interestingly, NGF binds its high affinity receptor, TrkA, in a position analogous to the location of the cavities in IL-17F. Figure 6B and C shows IL-17F and NGF in the same orientation, highlighting the locations of the cavities and the TrkA-binding sites (expected to be utilized by all neurotrophin/Trk complexes; Wiesmann et al., 1999). The known structures of neurotrophin homodimers (NGF, NT3, NT4) also have an indentation on their surfaces at this position but it is much smaller than the cavity in IL-17F (McDonald et al., 1991; Butte et al., 1998; Robinson et al., 1999). Trk family members are receptor tyrosine kinases that interact with neurotrophins via their membrane-proximal extracellular Ig-like domain. While the structures of IL-17R or IL-17Rh1 are not expected to contain an Ig-like fold, IL-17 ligands and neurotrophins could employ similar regions on their surfaces to bind their receptor.
Neurotrophins not only bind specific Trk receptors, but also can bind simultaneously to p75NTR, a second receptor common to all neurotrophins. p75NTR binds its neurotrophin ligands via a cystine-rich ECD that is expected to resemble the structures of TNF receptor 1 (TNFR1) or death receptor 5 (Banner et al., 1993; Hymowitz et al., 1999; Mongkolsapaya et al., 1999). A model of the NGF–p75NTR interaction has been proposed based on mutagenesis data (Wiesmann and de Vos, 2001) and suggests that the loops at either end of the ligand dimer as well as the flat surface on each protomer interact with p75NTR. The sequences of IL-17R and IL-17Rh1 do not resemble p75NTR, hence the proteins are not expected to adopt a TNFR1-like fold. However, given the similarity in IL-17 and neurotrophin folds, it is reasonable to consider the possibility of a second receptor component for IL-17s, analogous to the neurotrophin system.
The protein späztle has also been suggested to adopt a neurotrophin fold (Mizuguchi et al., 1998) and has been shown genetically (although not by direct binding experiments) to be a ligand for the Drosophila Toll receptor (Morisato and Anderson, 1994). Interestingly, IL-17A signals through NF-κB in a pathway similar to that used by IL-1 and Toll receptors, which share a common fold for their intracellular domains (although their ECDs are very different). Thus, either the intracellular or extracellular domains of IL-17 receptors, including other as yet unknown components of the signaling complex, may structurally resemble portions of these receptors. We propose that the assembly of the signaling complex, regardless of the fold of the receptor component(s) involved, is analogous to that for the neurotrophins, and specifically, that it will utilize ligand–receptor interactions involving the deep cavities in the sides of the IL-17 dimer.
Materials and methods
Protein expression and purification
An IL-17F cDNA clone was isolated from a human multiple tissue cDNA library and sequenced in its entirety. Essentially the same strategy was used to express and purify human IL-17F, IL-17A, IL-17E, IL-17R ECD (residues 1–288) and IL-17Rh1 ECD (residues 1–275). DNA containing the coding region for each protein of interest was first amplified by PCR, then subcloned into pET15b (Novagen) sites in order to introduce an N-terminal His tag and thrombin cleavage site. After another PCR step, the coding region was subcloned into the baculovirus transfer vector pAcGP67B (PharMingen), which was then co-transfected with BaculoGold DNA (PharMingen) into Sf9 cells, and recombinant virus was isolated and amplified in Sf9 cells.
For protein production, Hi5 cells were infected with amplified baculovirus. After 3 days in culture at 27°C, the medium was harvested by centrifugation. 50 mM Tris pH 7.5, 1 mM NiCl2, 5 mM CaCl2, 1 µM phenylmethylsulfonyl fluoride and 0.01% NaN3 were added and the pH was adjusted to 7.0. The medium was filtered and loaded onto a Ni-NTA (Qiagen) column. The column was washed with 50 mM sodium phosphate pH 7.0, 500 mM NaCl, 10 mM imidazole, then eluted with 250 mM imidazole in the same buffer. Fractions containing the protein of interest were pooled and dialyzed into phosphate-buffered saline (PBS) pH 6.5, together with 1 U/mg thrombin (Calbiochem) overnight at 4°C. The protein sample was then concentrated and the thrombin and His tag were removed by purification over a Superdex-75 column in 50 mM sodium phosphate pH 6.0, 500 mM NaCl. IL-17A, IL-17E and IL-17F all migrated as dimers, whereas IL-17R and IL-17Rh1 migrated as monomers on this column. IL-17A, IL-17E and IL-17F exist predominantly as disulfide-bonded dimers as shown by comparison of reducing and non-reducing SDS–PAGE. Relevant fractions were pooled and dialyzed into 50 mM sodium phosphate pH 6.0, 150 mM NaCl. For crystallization of IL-17F, the protein was instead dialyzed into 25 mM Bis–Tris propane pH 6.0, 100 mM NaCl and concentrated to 10 mg/ml. N-terminal sequencing confirmed the identities of all purified proteins and indicated that they each have the expected additional four amino acids (GSHM) at the N-terminus introduced by the vector’s cloning site. Mass spectrometry shows evidence of glycosylation of all proteins, with IL-17F containing ∼3 kDa carbohydrate per dimer.
Expression in activated T cells
Human peripheral blood mononuclear cells were isolated from blood using Lymphocyte Separation Medium (ICN/Cappel, Aurora, OH). Monocytes were depleted by plastic adhesion, after which CD4+ and CD8+ lymphocyte subpopulations were enriched by positive selection using MACS (Miltenyi Biotec GmbH, Germany). The purity of these cells was confirmed by FACS analysis. The enriched populations were cultured for 4 h in RPMI medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine and 1× penicillin/streptomycin, in both the presence and absence of a PI mixture of 5 ng/ml phorbol 12-myristate 13-acetate (PMA) and 3 µg/ml ionomycin. Total RNA samples were prepared using Qiagen RNeasy Mini or Midi kits. QIAshredder columns (Qiagen) were used to homogenize the cellular lysate. DNA was removed by DNase I digestion, both on the RNeasy column (Qiagen) and following elution (Ambion Inc., Austin, TX). IL-17F transcript level was measured by real-time quantitative RT–PCR, using primers and probes for 18S ribosomal RNA as a reference (PE Biosystems, Foster City, CA). Taqman reagents were specific for IL-17F and did not cross-react with IL-17A cDNA.
Articular cartilage explants
Human IL-17A and IL-1α (R&D Systems) were resuspended in buffer [PBS with 0.1% bovine serum albumin (BSA)] prior to use. The metacarpo-phalangeal joint of 4- to 6-month-old female pigs was aseptically opened, and articular cartilage was dissected free of the underlying bone. The cartilage was pooled, minced, washed and cultured in bulk for at least 24 h in a humidified atmosphere of 95% air, 5% CO2 in serum-free low glucose 50:50 Dulbecco’s modified Eagle’s medium:F12 media with 0.1% BSA, 100 U/ml penicillin/streptomycin (Gibco), 2 mM l-glutamine, 1× glycine/hypoxathine/thymidine, 0.1 mM MEM sodium pyruvate (Gibco), 20 µg/ml gentamicin (Gibco), 1.25 mg/l ampho tericin B, 10 µg/ml transferrin and 5 µg/ml vitamin E. Approximately 50 mg of articular cartilage were aliquoted into Micronics tubes and incubated for at least 24 h in the above media before changing to media without transferrin and vitamin E. Test proteins were then added. Media were harvested and changed at various time points (0, 24, 48, 72 h). Human knee articular cartilage, received from the National Disease Research Interchange (Philadelphia, PA), was cultured and treated in explants as above for porcine cartilage.
To measure proteoglycan breakdown, media harvested at various time points were assayed for amount of proteoglycans using the 1,9-dimethylmethylene blue (DMB) colorimetric assay (Farndale et al., 1986). Chondroitin sulfate (Sigma) ranging from 0.0 to 5.0 µg was used to make the standard curve. To measure effects on proteoglycan synthesis, [35S]sulfate (to a final concentration of 10 µCi/ml) (ICN Radiochemicals, Irvine, CA) was added to the cartilage explants at 48 h. After an overnight incubation at 37°C, media were saved for measurements of proteoglycan content. Cartilage pieces were washed twice using explant media. Digestion buffer containing 10 mM EDTA pH 8.0, 0.1 M sodium phosphate pH 6.5 and 1 mg/ml proteinase K (Gibco-BRL) was added to each tube and incubated overnight in a 50°C water bath. The digest supernatant was mixed with an equal amount of 10% w/v cetylpyridinium chloride (Sigma). Samples were spun at 1000 g for 15 min. The supernatant was removed, and 500 µl of formic acid (Sigma) were added to the samples to dissolve the precipitate. Solubilized pellets were transferred to scintillation vials containing 10 ml of scintillation fluid (ICN), and samples were read in a scintillation counter. Importantly, our recombinant IL-17A (prepared identically to IL-17F) had the same activity as IL-17A obtained commercially, indicating that the addition of amino acids at the N-terminus does not disrupt IL-17A function (not shown), and suggesting that the activity measured for our recombinant IL-17F represents native IL-17F activity.
Cytokine ELISAs
Conditioned media from explant cultures at 48 h were diluted 15-fold (porcine IL-6) or 150-fold (human IL-6) and used for assays. IL-8 and G-CSF production from cultured fibroblasts was determined as recommended by the assay manufacturer (R&D Systems).
Binding measurements
The kinetics and affinity of IL-17A, IL-17E or IL-17F binding to IL-17R or IL-17Rh1 were determined by SPR measurements on a Pharmacia BIAcore 1000 instrument (Pharmacia Biosensor, Piscataway, NJ). IL-17 ligand or receptor was immobilized onto a flow cell of a CM5 sensor chip covalently through primary amines according to the manufacturer’s instruction. An immobilization level of ∼500 resonance units (RU) was obtained for IL-17R, IL-17Rh1 and IL-17F, whereas IL-17A and IL-17E immobilization levels were 1200 and 1500 RU, respectively. After blocking unreacted sites with ethanolamine, binding measurements were performed using a flow rate of 25 µl/min. Sensorgrams were obtained for a series of six 2-fold serially diluted protein solutions. The highest concentration used was 1000 or 500 nM protein and the solutions were prepared in the running buffer, PBS containing 0.05% Tween-20. The sensor chip surface was regenerated between binding cycles by injection of a 25 µl aliquot of 0.1 M acetic acid, 0.2 M NaCl pH 3 to elute non-covalently bound protein. A strong signal was obtained for IL-17R binding to immobilized IL-17A. However, when IL-17R was immobilized, only a weak signal was obtained for IL-17A binding, suggesting that receptor immobilization inactivates the binding site. Sensorgrams were evaluated according to a 1:1 binding model by non-linear regression analysis using software supplied by the manufacturer. The association kinetics for the IL-17E–IL-17Rh1 interactions were not well described by the 1:1 binding model, suggesting either heterogeneity in the proteins or a more complicated binding mechanism. Nonetheless, the parameters obtained from the 1:1 binding model are reported in Table I. In separate experiments to measure competition between IL-17 variants for binding receptors, a fixed concentration of receptor was incubated with a varied concentration of IL-17 protein followed by injection of this mixture onto a flow cell having immobilized IL-17 protein. The amount of bound receptor was determined from the resonance signal obtained after completion of the association phase.
Crystallography
IL-17F crystallized as hexagonal plates in hanging drops over a well solution containing 1.0 M lithium sulfate, 0.5 M ammonium sulfate, 1% ethanol and 100 mM sodium citrate pH 5.6 at 19°C. Crystals were harvested into an artificial mother liquor consisting of the well solution without ethanol. Prior to data collection, crystals were immersed in artificial mother liquor with 20% glycerol and flash-cooled in liquid nitrogen. Initial data were collected on an in-house rotating anode generator with CuKα radiation and the space group was found to be P61 or P65, with two dimers in the asymmetric unit. For phasing, crystals were derivatized by soaking 6 h in artificial mother liquor supplemented with 2 mM thimerosal. A native data set and a three-wavelength Hg MAD experiment were collected at beam line 9-2 at the Stanford Synchrotron Radiation Laboratory. The data sets were processed using the programs in the HKL package (Otwinowski and Minor, 1997). Structure determination was carried out using the CCP4 suite of programs (CCP4, 1994). Patterson maps indicated the presence of several well-ordered Hg atoms whose locations were determined using the program Rantan. Phase refinement was carried out with MLPHARE. Examination of DM-modified maps indicated that the space group was P65, and revealed the non-crystallographic symmetry (NCS) operators. Each protomer bound a single thimerosal at an equivalent, NCS-related site.
The initial structure was built into a 4-fold NCS-averaged and solvent-flattened experimental map and was refined using the programs REFMAC_4.0 (CCP4, 1994) and X-PLOR (Brünger, 1992) as modified by Molecular Simulations, Inc. Reflections sequestered for calculating the free R-value were chosen in thin resolution shells. A maximum-likelihood target function, an overall anisotropic correction and a real-space bulk-solvent correction were used during positional refinement, simulated annealing and isotropic temperature factor refinement. Initial refinement was carried out against the 2.65 Å remote data set, but disorder around the Hg sites proved difficult to model so final refinement was carried out against the 2.85 Å native data set using the same set of free R reflections. A Ramachandran plot from Procheck (Laskowski et al., 1993) shows that 90% of all non-glycine, non-proline residues are in the most favored regions, 9.3% in the additionally allowed regions, 0.7% (three residues) in the generously allowed regions, with no residues in the disallowed regions. Data collection and refinement statistics are shown in Table II. The programs areaimol and resarea (CCP4, 1994) were used for accessible surface area calculations. The programs Molscript (Kraulis, 1991), Raster3D (Merrit and Murphy, 1994), Insight97 (Molecular Simulations, Inc.) and Grasp (Nicholls et al., 1991) were used for analysis and to make figures. The IL-17F coordinates have been deposited in the Protein Data Bank under accession code 1JPY.
Acknowledgments
Acknowledgements
We thank F.Vajdos, C.Eigenbrot, J.Stamos, M.Ultsch and the staff at SSRL beamline 9-2 for assistance with data collection; F.Vajdos for advice on MAD phasing; N.Skelton for threading analyses of IL-17 sequences; J.Bourell for mass spectrometry; and the protein sequencing group.
References
- Arend W.P. and Dayer,J.-M. (1990) Cytokines and cytokine inhibitors or antagonists in rheumatoid arthritis. Arthritis Rheum., 33, 305–315. [DOI] [PubMed] [Google Scholar]
- Awane M., Andres,P.G., Li,D.J. and Reinecker,H.-C. (1999) NF-κB-inducing kinase is a common mediator of IL-17-, TNF-α-, and IL-1β-induced chemokine promoter activation in intestinal epithelial cells. J. Immunol., 162, 5337–5344. [PubMed] [Google Scholar]
- Banner D.W., D’Arcy,A., Janes,W., Gentz,R., Schoenfeld,H.-J., Broger,C., Loetscher,H. and Lesslauer,W. (1993) Crystal structure of the soluble human 55 kd TNF receptor–human TNFβ complex: implications for TNF receptor activation. Cell, 73, 431–445. [DOI] [PubMed] [Google Scholar]
- Bowie A. and O’Neill,L.A.J. (2000) The interleukin-1 receptor/Toll-like receptor superfamily: signal generators for pro-inflammatory interleukins and microbial products. J. Leukoc. Biol., 67, 508–514. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR Manual, Version 3.1. Yale University Press, New Haven, CT.
- Butte M.J., Hwang,P.K., Mobley,W.C. and Fletterick,R.J. (1998) Crystal structure of neurotrophin-3 homodimer shows distinct regions are used to bind its receptors. Biochemistry, 37, 16846–16852. [DOI] [PubMed] [Google Scholar]
- Cai L., Yin,J., Starovasnik,M.A., Hogue,D., Hillan,K.J., Mort,J.S. and Filvaroff,E.H. (2001) Pathways by which interleukin 17 induces articular cartilage breakdown in vitro and in vivo. Cytokine, in press. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chabaud M., Fossiez,F., Taupin,J.-L. and Miossec,P. (1998) Enhancing effect of IL-17 on IL-1-induced IL-6 and leukemia inhibitory factor production by rheumatoid arthritis synoviocytes and its regulation by Th2 cytokines. J. Immunol., 161, 409–414. [PubMed] [Google Scholar]
- Chabaud M., Durand,J.M., Buchs,N., Fossiez,F., Page,G., Frappart,L. and Miossec,P. (1999) Human interleukin-17: a T-cell derived proinflammatory cytokine produced by the rheumatoid synovium. Arthritis Rheum., 42, 963–970. [DOI] [PubMed] [Google Scholar]
- Daun J.M. and Fenton,M.J. (2000) Interleukin-1/Toll receptor family members: receptor structure and signal transduction pathways. J. Interferon Cytokine Res., 20, 843–855. [DOI] [PubMed] [Google Scholar]
- Farndale R.W., Buttle,D.J. and Barrett,A.J. (1986) Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta, 883, 173–177. [DOI] [PubMed] [Google Scholar]
- Fossiez F. et al. (1996) T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J. Exp. Med., 183, 2593–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymowitz S.G., Christinger,H.W., Fuh,G., Ultsch,M., O’Connell,M., Kelley,R.F., Ashkenazi,A. and de Vos,A.M. (1999) Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with Death Receptor 5. Mol. Cell, 4, 563–571. [DOI] [PubMed] [Google Scholar]
- Jacobs K., McCoy,J.M., Kelleher,K. and Carlin,M. (1997) DNA sequences and secreted proteins encoded thereby. Patent application: WO 97/07198-A2.
- Jovanovic D.V., Di Battista,J.A., Martel-Pelletier,J., Jolicoeur,F.C., He,Y., Zhang,M., Mineau,F. and Pelletier,J.-P. (1998) IL-17 stimulates the production and expression of proinflammatory cytokines, IL-β and TNF-α by human macrophages. J. Immunol., 160, 3513–3521. [PubMed] [Google Scholar]
- Kingsley G.H. and Panayi,G.S. (1997) Joint destruction in rheumatoid arthritis: biological bases. Clin. Exp. Rheumatol., 15, S3–S14. [PubMed] [Google Scholar]
- Kong Y.-Y. et al. (1999) Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature, 402, 304–309. [DOI] [PubMed] [Google Scholar]
- Kotake S. et al. (1999) IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J. Clin. Invest., 103, 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lebecque S., Fossiez,F. and Bates,E. (2001a) IL-17. In Oppenheim,J.J. and Feldmann,M. (eds), Cytokine Reference, A Compendium of Cytokines and Other Mediators of Host Defense. Academic Press, San Diego, CA, pp. 241–250.
- Lebecque S., Fossiez,F. and Bates,E. (2001b) IL-17 receptor. In Oppenheim,J.J. and Feldmann,M. (eds), Cytokine Reference, A Compendium of Cytokines and Other Mediators of Host Defense. Academic Press, San Diego, CA, pp. 1541–1546.
- Lee J. et al. (2001) IL-17E, a novel proinflammatory ligand for the IL-17 receptor homolog IL-17Rh1. J. Biol. Chem., 276, 1660–1664. [DOI] [PubMed] [Google Scholar]
- Li H., Chen,J., Huang,A., Stinson,J., Heldens,S., Foster,J., Dowd,P., Gurney,A.L. and Wood,W.I. (2000) Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc. Natl Acad. Sci. USA, 97, 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindén A., Hoshino,H. and Laan,M. (2000) Airway neutrophils and interleukin-17. Eur. Respir. J., 15, 973–977. [DOI] [PubMed] [Google Scholar]
- Matusevicius D., Kivisakk,P., He,B., Kostulas,N., Ozenci,V., Fredrikson,S. and Link,H. (1999) Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Multiple Sclerosis, 5, 101–104. [DOI] [PubMed] [Google Scholar]
- McDonald N.Q. and Hendrickson,W.A. (1993) A structural family of growth factors containing a cystine knot motif. Cell, 73, 421–424. [DOI] [PubMed] [Google Scholar]
- McDonald N.Q., Lapatto,R., Murray-Rust,J., Gunning,J., Wlodawer,A. and Blundell,T.L. (1991) New protein fold revealed by a 2.3-Å resolution crystal structure of nerve growth factor. Nature, 354, 411–414. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Miossec P. (2000) Are T cells in rheumatoid synovium aggressors or bystanders? Curr. Opin. Rheumatol., 12, 181–185. [DOI] [PubMed] [Google Scholar]
- Mizuguchi K., Parker,J.S., Blundell,T.L. and Gay,N.L. (1998) Getting knotted: a model for the structure and activation of spätzle. Trends Biochem. Sci., 23, 239–242. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J., Grimes,J.M., Chen,N., Xu,X.-N., Stuart,D.I., Jones,E.Y. and Screaton,G.R. (1999) Structure of the TRAIL–DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nature Struct. Biol., 6, 1048–1053. [DOI] [PubMed] [Google Scholar]
- Morisato D. and Anderson,K.V. (1994) The spätzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell, 76, 677–688. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 176, 307–326. [DOI] [PubMed] [Google Scholar]
- Robinson R.C., Radziejewski,C., Spraggon,G., Greenwald,J., Kostura,M.R., Burtnick,L.D., Stuart,D.I., Choe,S. and Jones,E.Y. (1999) The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site. Protein Sci., 8, 2589–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwandner R., Yamaguchi,K. and Cao,Z. (2000) Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J. Exp. Med., 191, 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Barak T., Quach,J. and Lotz,M. (1998) Interleukin-17-induced gene expression in articular chondrocytes is associated with activation of mitogen-activated protein kinases and NF-κB. J. Biol. Chem., 273, 27467–27473. [DOI] [PubMed] [Google Scholar]
- Shi Y. et al. (2000) A novel cytokine receptor–ligand pair. J. Biol. Chem., 275, 19167–19176. [DOI] [PubMed] [Google Scholar]
- Spriggs M.K. (1997) Interleukin-17 and its receptor. J. Clin. Immunol., 17, 366–369. [DOI] [PubMed] [Google Scholar]
- Teunissen M.B., Koomen,C.W., de Waal Malefyt,R., Wierenga,E.A. and Bos,J.D. (1998) Interleukin-17 and interferon-γ synergize in the enhancement of proinflammatory cytokine production by human keratinocytes. J. Invest. Dermatol., 111, 645–649. [DOI] [PubMed] [Google Scholar]
- Van Kooten C. et al. (1998) Interleukin-17 activates human renal epithelial cells in vitro and is expressed during renal allograft rejection. J. Am. Soc. Nephrol., 9, 1526–1534. [DOI] [PubMed] [Google Scholar]
- Wiesmann C. and de Vos,A.M. (2001) Nerve growth factor: structure and function. Cell. Mol. Life Sci., 58, 748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C., Ultsch,M.H., Bass,S.H. and de Vos,A.M. (1999) Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature, 401, 184–188. [DOI] [PubMed] [Google Scholar]
- Yao Z., Fanslow,W.C., Seldin,M.F., Rousseau,A.-M., Painter,S.L., Comeau,M.R., Cohen,J.I. and Spriggs,M.K. (1995a) Herpesvirus saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity, 3, 811–821. [DOI] [PubMed] [Google Scholar]
- Yao Z., Painter,S.L., Fanslow,W.C., Ulrich,D., Macduff,B.M., Spriggs,M.K. and Armitage,R.J. (1995b) Human IL-17: a novel cytokine derived from T cells. J. Immunol., 155, 5483–5486. [PubMed] [Google Scholar]
- Yao Z., Spriggs,M.K., Derry,J.M.J., Strockbine,L., Park,L.S., VandenBos,T., Zappone,J., Painter,S.L. and Armitage,R.J. (1997) Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine, 9, 794–800. [DOI] [PubMed] [Google Scholar]
- Ziolkowska M., Koc,A., Luszczykiewicz,G., Ksiezopolska-Pietrzak,K., Klimczak,W., Chwalinska-Sadowska,H. and Maslinski,W. (2000) High levels of IL-17 in rheumatoid arthritis patients: IL-15 triggers in vitro IL-17 production via cyclosporin A-sensitive mechanism. J. Immunol., 164, 2832–2838. [DOI] [PubMed] [Google Scholar]