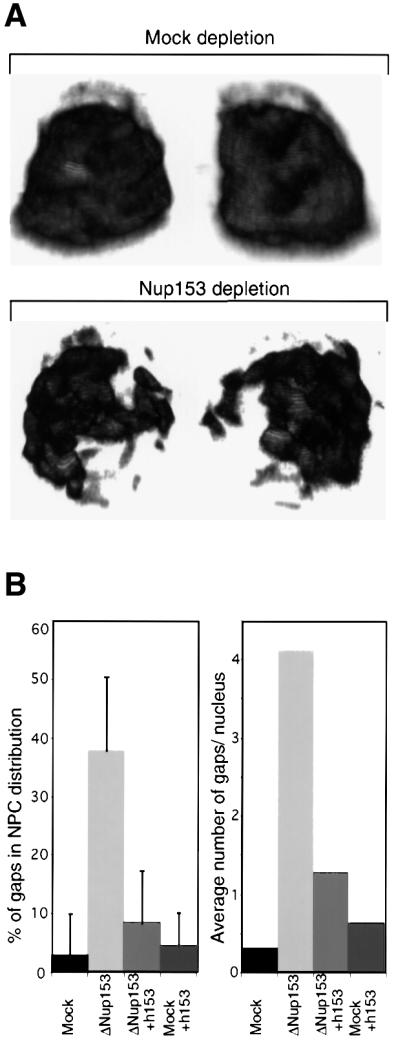

Fig. 3. Clustering of Nup153-deficient NPCs. Nuclei were assembled in vitro from mock-depleted or Nup153-depleted Xenopus egg cytosol and NPC distribution analysed using confocal immunofluorescence. (A) 3D reconstruction of confocal sections through a wild-type (Mock depletion) and Nup153-deficient (Nup153 depletion) nucleus stained with anti-Nup214/CAN primary and Alexa-488 conjugated secondary antibodies. Left hand images are rotated 180° as compared with right hand images. (B) Quantitation of NPC clustering. Nuclei were assembled from mock or Nup153-depleted (ΔNup153) Xenopus egg extracts in the presence (+h153) or absence of recombinant hNup153, and immunostained with mAb414 and Alexa-488 conjugated secondary antibody. Mid-plane confocal images were recorded and NEs were analysed for discontinuities. Both the percentage of NE length without nucleoporin staining (left panel) and the number of nucleoporin gaps in the nucleoporin staining per nucleus (right panel) are represented. Bars represent standard deviations. (C) Representative immunofluorescence images of mid-plane confocal sections of synthetic nuclei analysed in (B). Nucleoporin staining is in red, chromatin staining (DAPI) in blue.
