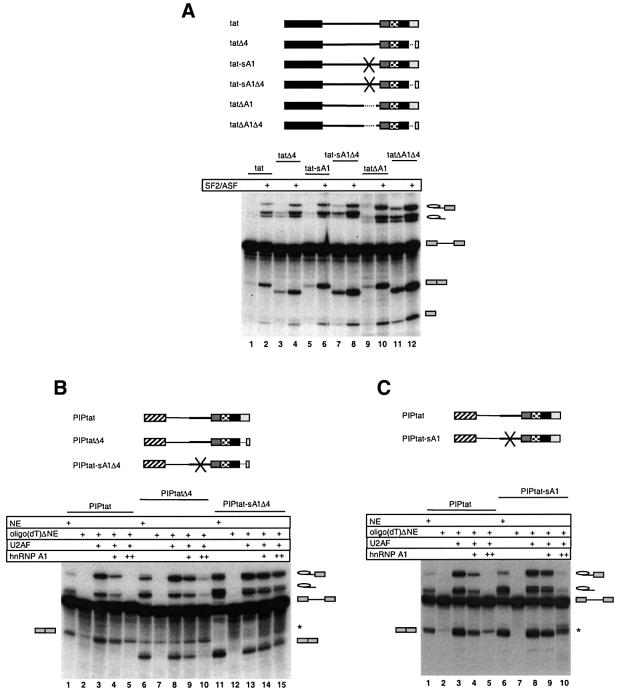
Fig. 6. In vitro splicing analysis determining the effect of ISS and ESS3 mutations. (A) Splicing of tat pre-mRNA in NE in the absence and presence of 32 ng/µl recombinant SF2/ASF (lanes 1 and 2) as compared with tat-derived pre-mRNAs with deleted ESS3 (tatΔ4; lanes 3 and 4), scrambled ISS (tat-sA1; lanes 5 and 6), deleted ISS (tatΔA1; lanes 9 and 10), scrambled ISS and deleted ESS3 (tat-sA1Δ4; lanes 7 and 8), and deleted ISS and ESS3 (tatΔA1Δ4; lanes 11 and 12). (B) Splicing of the chimeric PIPtat pre-mRNA (lanes 1–5), as compared with PIPtat-derived pre-mRNAs with deleted ESS3 (PIPtatΔ4; lanes 6–10), and scrambled ISS and deleted ESS3 (PIPtat-sA1Δ4; lanes 11–15). (C) Splicing of PIPtat pre-mRNA (lanes 1–5) and PIPtat with scrambled ISS (PIPtat-sA1; lanes 6–10). The source of NE was: non-depleted NE, oligo(dT)ΔNE or oligo(dT)ΔNE supplemented with U2AF alone or together with 12.5 ng/µl (+) or 37.5 ng/µl (++) (final concentration) GST–hnRNP A1 as indicated above the lanes. The asterisk indicates a degradation product that co-migrates with the exon–exon product of PIPtat. The identities of the splicing products are indicated.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.