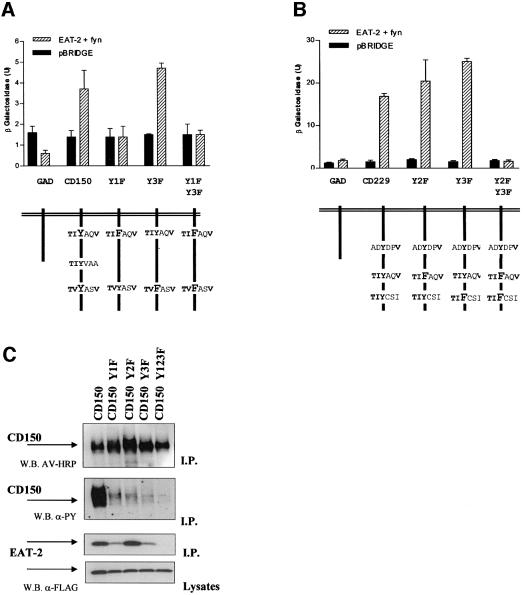
Fig. 7. Analysis of EAT-2 binding to Y–F CD150 and CD229 mutant receptors. (A) EAT-2 binding to CD150 Y–F mutants in a direct interaction hybrid system. Hybrid system direct interaction analysis of the binding between EAT-2 in pBRIDGE-Fyn420,531Y–F (dashed bars) and different cD150 Y–F mutations in pGAD424. As control, vector pBRIDGE was used (solid bars). In particular, mutants CD150 Y1F (Tyr281 to phenylalanine), Y3F (Tyr327 to phenylalanine) or Y13F (Tyr281 and Tyr327 to phenylalanine) were used in this study. The lower part of the figures depicts the mutations in the CD150 cytoplasmic tail. (B) EAT-2 binding to CD229 Y–F mutants in a direct interaction hybrid system. Hybrid system direct interaction analysis of the binding between EAT-2 in pBRIDGE-Fyn420,531Y–F (dashed bars) and different cD229 Y–F mutations in pGAD424. As control, vector pBRIDGE was used (solid bars). In particular, mutants CD229 Y2F (Tyr558 to phenylalanine), Y3F (Tyr581 to phenylalanine) and Y23F (Tyr558 and Tyr581 to phenylalanine) were used in this study. The lower part of the figure depicts the mutations in the CD229 cytoplasmic tail. (C) EAT-2 binding to CD150 Y–F mutants in an in vivo system. Full-length wild-type CD150, CD150 Y1F (Tyr281 to phenylalanine), CD150 Y2F (Tyr307 to phenylalanine), CD150 Y3F (Tyr327 to phenylalanine) and CD150 Y123F (Tyr281, Tyr307 and Tyr327 to phenylalanine) were co-transfected with EAT-2 and fyn in a COS-7 cell system. Mutant cD150 cDNAs used in the transfections are indicated above each set of panels. Immunoprecipitates (I.P.) were made with an anti-CD150 antibody and analyzed by western blotting. Anti-phosphotyrosine = W.B. α-PY; avidin = W.B. AV-HRP; EAT-2 = W.B. α-FLAG. The bottom panel shows the lysates of the same cell extracts used for the precipitation.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.