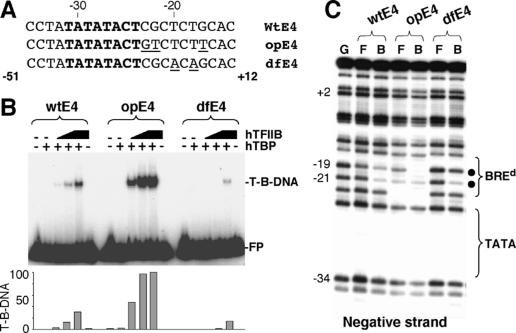
Figure 2.
The BREd modulates assembly of a TFIIB–TBP–promoter complex. (A) The wild-type AdE4 (wtE4) sequence is shown along with derivatives containing a BREd sequence that has either been optimized (opE4) or rendered low affinity (dfE4). (B) Bandshift assay comparing the assembly of a TFIIB–TBP–promoter complex with the wild-type AdE4 promoter (wtE4) and optimized (opE4) or defective (dfE4) derivatives. Free probe (FP) and TFIIB–TBP–DNA complex (T-B-DNA) are indicated. TBP was 50 ng and TFIIB was 1.25, 5, and 20 ng. (C) Methylation interference assay analyzing the wild-type AdE4 promoter (wtE4) and optimized (opE4) or defective (dfE4) derivatives. (G) G-track generated by cleavage of the partially methylated wild-type AdE4 promoter with piperidine; (F) free probe; (B) TFIIB–TBP-bound DNA. Sites of methylation interference are denoted by a solid circle. The region of the TATA box and BREd are indicated. The numbers at left represent the position relative to the transcription start site.