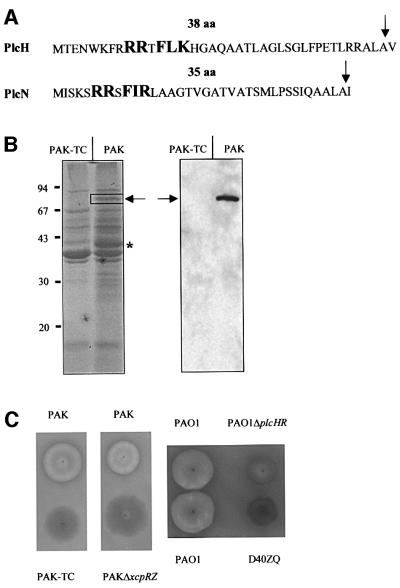
Fig. 3. N-terminal signal sequences of PlcH and PlcN, protein profiles of the supernatants and haemolytic activities of the wild type and the tatC mutant (PAK-TC). (A) The twin-arginine motif R-R-x-F-(I/L)-(K/R) is represented in bold and upper case letters. The number of residues (aa) in each signal peptide is indicated. The signal peptide cleavage site is indicated by an arrow. (B) Proteins from culture supernatants of PAK and PAK-TC strains equivalent to 1 absorption unit of cells were separated on a denaturing SDS–gel containing 11% acrylamide and stained with Coomassie Blue (left panel), or immunoblotted and revealed by using monoclonal antibodies directed against PlcH (right panel). The expected migration position of PlcH and PlcN is indicated by an arrow. The bands analysed by mass spectrometry and missing in the tatC mutant are boxed (PlcN) or indicated with an asterisk (GlpQ). The positions of molecular weight markers (kDa) are indicated on the left. (C) Haemolytic activity on blood agar plates of PAK or PAO1, and the derivative xcpR-Z or plcHR and xcpZ-Q (D40ZQ) mutant strains, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.